A Career, in Sequences

Claire M. Fraser speaks with ease, humor, and pride about the people and projects that have contributed to the success of The Institute for Genomic Research (TIGR) in Rockville, Maryland. She’s certainly had plenty of practice, having given multiple stories and interviews to local and national science media since she took over leadership of TIGR from founding President Craig Venter in 1998. She is often asked to compare her management style with that of the outspoken Venter, also her husband of over twenty years, and to comment on the challenges of working closely with her spouse. She does not shy away from such questions, nor is she in any way reticent about her own accomplishments as a biomedical researcher and now administrator, but she is much more likely to concentrate on the biological questions, as well as the “fun,” that have motivated her throughout her career. She seems still to be busy having fun, whether she is helping her faculty reach their goals, publicly representing the capabilities and needs of TIGR, or talking about the science of genomics, microbiology, and evolution.
MI: It’s not at all obvious that your career path, leading to your present position as President of TIGR, should have begun with a PhD in pharmacology. Why did you choose to go into pharmacology?
CF: Well, you might just ask, “Why graduate school?” to begin with, and that all sort of ties in. I was certain from the time I was a freshman in high school that I was going to have some sort of career in science. I grew up in a small town outside of Boston, and I didn’t know anything about scientific research—I didn’t know anybody who had chosen that career path. The only way I knew to go about getting into science was to go into medicine. As a college senior [at Rensselaer Polytechnic Institute, RPI], I went through the whole process of applying to medical school, and I started to go on interviews. But at the same time, I had had the opportunity, as a senior, to work in a lab doing a bona fide research project—not a canned laboratory exercise, and I just fell in love with it. I spent every free moment in my last year of college, instead of at a bar in the student union, having fun in the lab.
MI: It’s a common story.
CF: It was wonderful! And as I got closer to the end of my senior year, I had gotten to know a lot of the graduate students at RPI, and so all of a sudden, this alternative career path became known to me. At the very last minute, I made the decision that I was not going to go to medical school. But I had put so much time into thinking about where I was going to go to medical school that I didn’t really know where to begin to apply to graduate programs. I had been dating a guy who had graduated a year ahead of me, and he was working in Toronto. So, I started to think, “Since I’m starting from scratch and have to pick another school, maybe I can find one close to Toronto.” Buffalo was as close to Toronto as you could get without leaving the US—I shouldn’t even be confessing this!—but it’s exactly what happened. So, I picked up a Peterson’s guide and basically looked to see what was available at the State University of New York in Buffalo. I went through descriptions of all the research activities in the various departments in the medical school—I wanted to be in a graduate program in a medical school because I was still somewhat on the fence about career paths. Pharmacology sounded more interesting than anything else! I wish I could say that I had really been led by an interest in pharmacology, but at that point, I was just looking for something that caught my interest. In Buffalo. I’m sort of embarrassed to confess this.
MI: That’s a great story…
CF: It’s a pathetic story!
MI: …well, it shows that things can work out even if you don’t have the best plan.
CF: Things can work out. And there actually was some continuity between the research that I had been doing as an undergraduate and the research activities going on in the pharmacology department that I entered in Buffalo. My undergraduate research was in a microbiology lab, looking for bacteria from Lake George that could break down pesticides and herbicides. The department I entered in Buffalo had efforts in the areas of both pharmacology and toxicology, and there were a couple of microbiologists on the faculty.
MI: After entering the program in Buffalo, did you begin to identify yourself as a pharmacologist?
CF: Absolutely! Once I got into graduate school, I really started to focus. I started working on G protein–coupled receptors. A lot of my thesis work in graduate school depended on developing monoclonal antibodies against GPCRs and trying to develop immunoaffinity purification protocols to isolate and characterize receptors. I was starting to think about how you define receptors from a molecular point of reference, rather than just as black-box proteins that are in the cell membrane. One paper that caught my attention when I was in grad school was the Kohler-Milstein paper on monoclonal antibodies. That’s where we began to think about producing antibodies against various receptor types and using the specificity of monoclonal antibodies to map some of the differences. We were able to begin to think about experiments to uncover the molecular basis for the differences in specificity among receptor types. I was initially focusing just on the β1 - and β2 -adrenergic receptors that were very similar in terms of their properties.
MI: So, you really became a hard-core pharmacologist. Would you still think of yourself as an advocate for pharmacology?
CF: Absolutely. I think that one of the things that I find most intriguing from our studies of microbial genomes is that one of the largest classes of proteins that you see are transporters. I don’t really think of transporters as being all that different from receptors: both classes of protein are found on the surface of the cell; both classes engender a range of specificity of interactions with ligands; and the large families of transporter molecules that we see are very analogous to the receptor situation that I was working on over twenty years ago as a graduate student. Many bacterial transporters look like they’re likely sugar transporters, and our obvious goal is to uncover the functions and specificities of individual transporters. So, here’s a clear parallel with the type of receptor work that typifies pharmacological investigations.
MI: A couple of years after finishing your degree in Buffalo you moved your work in GPCRs to the NIH. What prompted that particular move?
CF: We went to NIH because NIH was really sort of like going to Disneyland, in a way, for investigators who were used to surviving in the extramural system, having to write grants. You go to the NIH, you get a very generous budget, you spend the money, and if you can show some productivity, you usually get even more funding the following year. It was a completely different system, and it was like a dream come true.
MI: And how long did the dream last?
CF: Oh, NIH was fabulous, and if that’s all there was for the rest of my career, I suspect I would have been pretty darned happy. I did eventually get tenure just before I left the NIH (in 1992). I was in the enviable situation of not having to write for grants. I had access to all sorts of resources within the intramural program that a lot of outside investigators don’t have access to. When we first moved to the NIH, that’s when we learned molecular biology. Before that, we, as well as a few other labs, had been trying to do large-scale purification of receptor proteins as a way to eventually clone the genes that encode these receptors. Once that happened, we began to do a lot of site-directed mutagenesis, mapping ligand binding sites and receptor–G protein interaction sites. I thought that was just great stuff, but I had always been interested in GPCRs as a family, and interested in the evolution of the families. As you know, these are proteins that in some cells are present in only a few hundred copies.
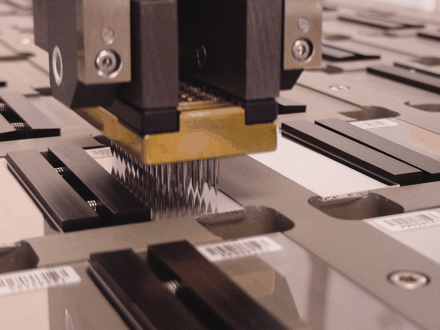
A TIGR robot “spots” microaray slides in experiments to determine the gene function in one of the Institute’s many projects.
What brought me out of NIH to TIGR in 1992 was the possibility of really thinking about approaching the evolutionary question in a big way. When we made the move, there were probably about thirty or so GPCRs that had been cloned and sequenced, but it was estimated that several hundreds to thousands of GPCRs existed, which has been verified now with genome sequence information. I was excited about using the EST [expressed sequence tag] approach as a way to find new receptor proteins. The EST method had been proven to be very robust, and we knew enough about these receptor proteins that we could identify signature sequences that could lead us to new receptors. So, that’s what I was really interested in doing: Finding new receptor proteins, doing some of the functional work to start to look at identifying potential ligands, and ultimately collecting this information and starting to build information on GPCRs as a large family. Once at TIGR, we ended up identifying probably another hundred or so new receptors in a fairly short amount of time through the EST approach.

TIGR’s core sequencing laboratory is one of the world’s largest and best.
The only problem with the GPCR work was that TIGR’s funding came from Human Genome Sciences, which was also founded at the same time as TIGR. Human Genome Sciences and its first partner, SmithKline Beecham, had access to our intellectual property. All of the GPCRs are potential drug targets, and so even though we had done such a good job of identifying them, we couldn’t publish. I found this to be incredibly frustrating, and I was feeling really sorry for myself, sensing that my receptor career had stalled.
MI: Did this situation havae any impact on your abrupt shift in research direction?
CF: No question about it. When I made the shift [to microbial genomics], I originally envisioned that it would be temporary and I would go back to receptor work, but I was also so excited about the possibilities of thinking about biology on a large scale, at a whole-genome level. While I was sitting around feeling really sorry for myself about losing control of the GPCR work, the Haemophilus project1 had started up here at TIGR, and I was somewhat involved in that, but more peripherally. But I was clearly aware of the excitement that was being generated as that project came to completion, and I did get in on some of the analysis work at the end because we wanted to put as many able minds as we could on trying to do this analysis. But thinking about it for the first time, it was a pretty daunting task.
I led the second microbial genome project at TIGR—the genome of Myoplasma genitalium , the simplest organism. We selected that as the second project because upon finishing the Haemophilus genome, we were surprised by finding such a large percentage of genes of unknown function. We had all been convinced, even while we were doing the analysis of Haemophilus , that we would be able to recognize all the genes and put them into pathways—that we’d see the circuit diagrams for Haemophilus and we’d understand it all. But all of a sudden that little fantasy quickly disappeared; thirty-something percent of the structural genes encoded proteins whose function could not be deciphered by sequence alone. So, we targeted Myoplasma genitalium as the second organism because its genome size had been estimated to be between 400 and 600 genes, and that was a good estimate—we found 470. We thought that certainly we were smart enough to figure out what 470 genes do, even if we couldn’t figure out the 1700+ genes in Haemophilus . But in Mycoplasma genitalium we found the same thirty-something percent of genes encoding proteins of unknown function. I remember that as being a critical point. We realized that we weren’t as smart as we thought we were!
I loved getting into the sequencing projects: I’d sit at a table with every biochemistry textbook, every molecular biology textbook, stacks of references about the specific organism, and really try to put the biology back together based on gazing at the genome sequence. Although there was this big part of the genome that we couldn’t understand, in every case there were enough examples where you could put together disparate pieces of information and all of a sudden come up with molecular explanations of something that might have been published ten years earlier. I found that so intellectually satisfying that I was really hooked. I’ve always been interested in the evolution of gene families, and now we were able to think about evolution of species.
MI: Your interests were still led by biological questions then? You didn’t have to become a technophile, concentrating on the new technology rather than on biology?
CF: No. Compared to many “genome people,” I’m less interested in the technology than I am in what you can do with the information that you get. And I could come up with a list of people in the genome field—and I don’t think there would be much disagreement—about whom I could say, “They’re more interested in the technology and pushing the technology development.” Because I came into the field in a somewhat roundabout way—sort of by accident, if you will—I’ve never been driven by the technology. Of course, technology is critically important. It’s what’s driven the field, and will continue to drive it in the future, but if somebody else wants to worry about the technology—great. I’d rather worry about how you’re going to apply data to answer biological questions.

The main building to TIGR’s eighteen-acre campus in Rockville, MD. has a bright California-style look.
MI: Have you been forced to confront technology to a degree that you didn’t want to?
CF: No, because what really has become clear about the genomics field is that it requires such a multidisciplinary approach that no single individual can do all of it. Even within TIGR, we have folks who are more interested in sequencing and sequencing technology, and we have other people who are interested in bioinformatics, developing new algorithms, developing new tools, and still other people who are more the users of that bioinformation. And so it’s been very easy for everyone to find a niche.
MI: Speaking of finding a niche, how have you adapted to life as a science administrator? Does administration give you a chance to look at these different—multidisciplinary—things in a satisfying way?
CF: I think it does. I’ve just had to come to grips with getting intellectual satisfaction from doing things outside of the lab. And now I really see my role as much more of a facilitator for the entire faculty here. And I think we’ve done extraordinarily well since I took over in 1998; the research program has grown tremendously. TIGR’s reputation has continued to grow. And I think I can take some credit for that. My role now gives me as much satisfaction as sitting with all the biochemistry books used to.
MI: Where do you get the wherewithal to be an administrator at your level?
CF: I have absolutely no idea! I have to tell you that I was scared to death when I took over. I figured, “Oh my god, I’m going to screw this up big-time.”
MI: Did you really?
CF: Yeah, I truly did. Because it wasn’t ever something I aspired to even secretly. It wasn’t something I was ever planning on. I was never one to think, “What do I do next to position myself to take on such a role in the future?” It was really thrust upon me unexpectedly. And I can tell you, and Craig will agree, we had some pretty serious shouting matches, with me yelling, “Why are you doing this? Why do you have to go sequence the human genome? Why do you have to leave TIGR? Everything is great and you’re just screwing it up!”
MI: So, why did you agree to take on the role of President of TIGR?
CF: Because we really had, I think, created a unique culture here.
MI: In what way?
CF: Well, we had an incredible opportunity to be able to set something up from nothing and really create an organization in what I think was our collective vision. And there really were some philosophical approaches—about how you approach science, how you hire people, how you build teams, what you place the value on—that you can’t be sure somebody coming in from the outside would necessarily see the same way. And you can’t place somebody else in charge and then say, “But wait a minute. You have to do things our way.” Craig and I have always been in agreement that you shouldn’t let titles of positions or educational backgrounds get in the way of letting people do what they want to do. I don’t want an administration that gets in the way of keeping our people from developing their full potential.
Footnotes
-
↵1
- © American Society for Pharmacology and Experimental Theraputics 2003



