Knockout Mouse Points to Second Form of Tryptophan Hydroxylase
- 1Department of Psychiatry,
- 2Departments of Pediatrics and Human Genetics; Committees on Clinical Pharmacology and Pharmacogenomics, Molecular Medicine, Genetics, The University of Chicago, Chicago, Illinois 60637
- EHC ed{at}yoda.bsd.uchicago.edu; fax 773-834-0505.
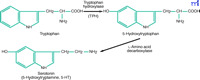
Tryptophan hydroxylase (TPH, EC 1.14.16.4), a tetrahydropterin-dependent amino acid hydroxylase, catalyzes the rate-limiting step in the synthesis of 5-hydroxytryptamine (5-HT, serotonin) from tryptophan (Figure 1⇓). The C-terminal regions of the three known tetrahydropterin-dependent amino acid hydroxylases, which also include phenylalanine hydroxylase and tyrosine hydroxylase, share a high degree of sequence similarity, suggesting that they arose from a common ancestral gene (1) . In the brain, 5-HT synthesis occurs primarily in the dorsal raphe nucleus (DRN). Serotonin serves as an intermediate in melatonin synthesis in the pineal gland (PG), where TPH is also expressed; however, TPH is not the rate-limiting enzyme in this pathway. In the periphery, 5-HT is produced by enterochromaffin cells in the gut.
Earlier observations reported contradictions between the levels of expression of Tph1 mRNA and protein, and complicated regulatory means of translational efficiency were invoked to explain much of the confusion. For example, TPH from neoplastic mouse mast cells and TPH extracted from rat brain were found to have different expression rates, regulation, and molecular mass (2) . Second, Tph1 mRNA is surprisingly scarce and heterogeneously distributed in the DRN (3) , so much so that PCR-based techniques are often necessary just to detect the transcripts (4) . Furthermore, Tph1 mRNA levels are ten- to fortyfold higher in the PG than in the DRN, but the amount of protein is comparable in each tissue (3, 4) . Intrigued by these findings, investigators identified two Tph mRNA species arising from different promoters and having different translational efficiency. The expression of the more efficiently translated isoform is increased in response to stress, but these findings still do not account for the observed differences in expression of TPH mRNA and TPH protein (4) .
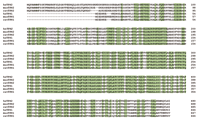
The TPH mRNA–protein puzzle took on new significance when Walther and colleagues demonstrated that a Tph1 knockout mouse failed to generate the expected phenotype: Tph1 -deficient mice continued to produce 5-HT in the brain, but had almost no detectable serotonin in the duodenum, and none in whole blood (5) . Walther and colleagues then found Tph2 with remarkable ease. Molecular biological techniques were necessary only for confirmation—cells transfected with Tph2 acquired tryptophan-hydroxylating activity (5) . Primarily utilizing bioinformatics, the authors found part of a Tph gene paralog on murine chromosome 12 in the Genbank database. Subsequently, they identified homologous genes in the rat and human (Figure 2⇓). Thus, the identification of a brain-specific, second tryptophan hydroxylase gene (Tph2 in mouse, TPH2 in human; also tentatively named neuronal TPH, NTPH ) may explain some earlier contradictions noted between the expression of Tph1 mRNA and protein (5) .
Importantly, Walther and colleagues found that the antibodies commonly used to identify TPH cross-reacted to both Tph1 and Tph2 gene products (5) . On the other hand, the 73% similarity at the mRNA level is unlikely to be confused in expression studies, suggesting that previous protein studies in DRN were likely describing TPH2 distribution while comparing it with Tph1 mRNA levels. Although Walther and colleagues did not test for TPH2 or TPH1 expression patterns in the human, a study on post-mortem human brain found similar patterns of TPH1 mRNA as in the rat PG and DRN (3) .
The discovery of Tph2 serves as the first step in clarifying the regulation of serotonin synthesis in the brain. The work by Walther et al. (5) also calls into question numerous studies that now appear to have tested the genetic relationship between a peripherally expressed enzyme and its function in central nervous system (CNS) function. Additionally, hypotheses attempting to link peripheral and central measures of serotoninergic activity should be reexamined.
Tryptophan hydroxylase was never thought to be a strong target for drug discovery because of its wide-ranging function; however, an isoform expressed primarily in the periphery and pineal gland may be a candidate for novel anti-emetic or gut-motility agents, especially given the success of ondansetron, a 5-HT3 receptor antagonist (6) . Conversely, the identification of brain-specific TPH2 might foster drug development in which the CNS form of TPH can be modulated with little or no peripheral effects. Investigators have studied the regulation of TPH, including the role of phosphorylation and interactions with the 14-3-3η protein (7) . Indeed, the regulation of the enzymes encoded by these two genes may differ significantly, offering additional regulatory proteins as putative drug targets.
The discovery of a second TPH gene alters the field of candidate genes and proteins for scientists seeking to understand the relationship between peripheral measures of 5-HT and disease. For example, since the initial report of hyperserotonemia in autism (8) , biological psychiatry has sought to use the serotonin system in the platelet as a model for the synapse (9) . Essentially, abnormalities in the platelet serotonin system are thought to reflect variations in expressed genes and proteins that are also important in synaptic transmission and regulation. The discovery of separate TPH genes responsible for peripheral–PG versus DRN 5-HT synthesis suggests that the synthetic portion of the system is less likely to be the source of important variation contributing to peripheral findings in CNS disease, because different TPH isozymes are responsible for 5-HT production in each location. Previous efforts to connect variation in TPH1 and TPH expression will need to be repeated with antibodies specific to each protein isozyme (10) .
The implications of a second TPH gene are exciting, presenting new opportunities to understand the interrelated serotonin system. A division of labor in 5-HT synthesis fundamentally changes our approach to peripheral models of CNS disease just as unexpected discoveries challenge us to rethink our paradigms. The identification of Tph2 by Walther et al. (5) reinforces the importance of data mining in the post-Human Genome Project era. We might be surprised just how easy it is to find large gene needles in database haystacks.
The tryptophan to serotonin two-step. The synthesis of serotonin from trytophan requires two enzymatic reactions. The first catalytic step involving the hydroxylation of tryptophan by TPH is rate limiting.
Alignment of the predicted amino-acid sequences o1 TPH1 and TPH2 from man, mouse and rat. Comparison of the sequences of TPH1 to TPH2 within each species reveals amino-acid identities of 72%, 68%, and 70% for man (hs), mouse (mus), and rat (rat), respectively. (Reprinted with permission from Walther et al. Synthesis of serotonin by a second tryptophan hydroxylase isoform. Science 299, 76 (Supplementary material) (2003). American Association for the Advancement of Science.)
Footnotes
-
Edwin H. Cook, Jr., MD , is a Professor of Psychiatry, Pediatrics, and Human Genetics and member of the Committees on Pharmacology and Pharmacogenomics, Molecular Medicine, and Genetics at the University of Chicago. He is the director of the Laboratory of Developmental Neuroscience. Jeremy Veenstra-VanderWeele, MD , is a Resident Physician in Psychiatry and a member of the Laboratory of Developmental Neuroscience at the University of Chicago, Pritzker School of Medicine.
- © American Society for Pharmacology and Experimental Theraputics 2003