Identification of TOR-interacting Proteins
- Kazuyoshi Yonezawa
- Biosignal Research Center, Kobe University, Kobe 657-8501, Japan, and CREST, Japan Science and Technology Corporation
The target-of-rapamycin (TOR) proteins are protein kinases that were first identified in Saccharomyces cerevisiae through mutants that conferred resistance to growth inhibition induced by the immunosuppressive macrolide rapamycin (1 , 2). In yeast, wild-type TOR1 and TOR2 control a variety of processes contributing to cell growth—in response to nitrogen availability—including
translational initiation and early G1 progression (3) , as well as the regulation of transcription, amino-acid uptake, cytoskeletal organization, and protein degradation through
autophagy (4). In mammalian cells, rapamycin blocks the phosphorylation of eukaryotic initiation factor 4E-binding protein 1 (4E-BP1) (5 , 6) and p70 S6 kinase (p70S6K) (7 , 8) by interfering with the function of mTOR [also known as FK506-binding protein (FKBP) 12-rapamycin–associated protein (FRAP),
or rapamycin- and FKBP-target 1 (RAFT1)] (9 , 10). Although mTOR can phosphorylate both these targets directly in vitro (11 –13) , the regulation of the kinase activity of mTOR in vivo remains incompletely understood. In addition, nutrients, especially
amino acids, which can regulate the phosphorylation of p70S6K and 4E-BP1, are necessary for insulin or mitogen regulation
(14 –19). Despite extensive efforts, how nutrients regulate the mTOR signaling pathway remains poorly understood.
The recent publications of several reports, that unveil a series of TOR-interacting proteins in yeast and mammalian cells,
have given new insights into the TOR signaling pathway. One of TOR-interacting protein is Raptor (regulatory associated protein
of mTOR) or its S. cerevisiae ortholog Kog1p, a highly conserved 150-kDa TOR-binding protein (20 –22). All Raptor orthologs contain a unique conserved region in their N-terminal half (raptor N-terminal conserved, also called
the RNC domain) followed by three HEAT (huntingtin, elongation factor 3, A subunit of protein phosphatase 2A and TOR1) repeats
and seven WD-40 repeats near the C terminus. Research on mammalian Raptor suggests that its association with mTOR promotes
the phosphorylation of downstream effectors in nutrient-stimulated cells (20 , 21). In concordance with these observations, the binding of TOR to Raptor or to Kog1p (22) is necessary for TOR signaling in vivo in Caenorhabditis elegans and S. cerevisiae (21 , 22).
Another characterized mTOR-interacting protein from S. cerevisiae Lst8p—its mammalian ortholog is mLST8/Gβ L (G protein β -subunit-like protein, pronounced “gable”)—a highly conserved 36-kDa
protein that consists almost entirely of seven WD-40 repeats with high sequence similarity to those found in the β subunits
of heterotrimeric G proteins (22 , 23). Loewith et al. reported that Lst8p interacts with Tor1p and Tor2p and showed that transiently expressed recombinant mTOR and mLST8 can
interact (22). Kim et al. (23) independently identified the same interacting protein and adopted the Gβ L name based on a previous report (24).
Regarding Raptor function, Sabatini and colleagues have also reported that the stability of the mTOR–Raptor complex is increased
when cells are amino acid– or energy-starved (20). The transition to this avid mTOR–Raptor complex correlates with the inhibition of mTOR-dependent signaling in cells and
with the repression of mTOR’s kinase activity in vitro, leading Sabatini’s group to suggest that Raptor negatively regulates
the kinase activity of mTOR. Very recently, Kim et al. observed that mLST8/Gβ L interacts constitutively with and activates the kinase domain of mTOR, and that mLST8/Gβ L is necessary
for mTOR to form a nutrient-sensitive interaction with Raptor (23). These authors favor a model in which the binding of Raptor to the complex of LST8/Gβ L and mTOR inhibits mLST8/Gβ L-mediated
activation of mTOR, and propose that the opposing effects on mTOR activity of the interactions mediated by mLST8/Gβ L and
Raptor provide a mechanism by which cellular conditions, such as nutrient levels, can positively and negatively regulate mTOR
signaling to the cell growth machinery (23).
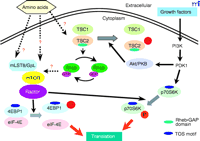
Figure 1.
A working model for the mTOR signaling pathways in mammals. Nutrients regulate mTOR signaling pathway and Raptor and mLST8/GβL are components of the mTOR signaling complex. Raptor serves
as a scaffolding protein that binds p70S6 kinase (p70S6K) and eukaryotic initiation factor 4E-binding protein 1 (4E-BP1) through
their TOR-signaling (TOS) motifs and Raptor facilitates their phosphorylation by mTOR. mLST8/GβL is another component of the
mTOR signaling complex. mLST8/GβL interacts constitutively with the kinase domain of mTOR. The right side of the figure outlines
a tentative pathway that links growth factor–dependent Akt/PKB activation through phosphatidylinositol-3’ kinase (PI3K) and
phosphoinositide-dependent kinase 1 (PDK1) to the stimulation of mTOR-dependent responses. The tuberous sclerosis complex
(TSC) proteins TSC1 and TSC2 serve as negative modulators of the mTOR pathway. The small GTPase Rheb (Ras homolog enriched
in brain) is a direct target of TSC2’s intrinsic GTPase-activating function. Rheb•GTP appears to be a positive modulator of
mTOR signaling. PKB, protein kinase B.
However, we have studied the same protein–protein interaction and have found no evidence for changes in Raptor–mTOR complex
stability when cells were shifted between nutrient-rich and nutrient-poor media (21). In addition, we observed that coprecipitation of Raptor with mTOR is essential for the phosphorylation of 4E-BP1 and p70S6K
(in immune-complex kinase assays) and that Raptor also binds to p70S6K and to 4E-BP1. Thus, we propose that Raptor participates
as an essential scaffolding protein for mTOR-catalyzed phosphorylation of 4E-BP1 and p70S6K (21). Obviously, important technical differences between our and Sabatini’s group—such as cell lysis conditions and studying transfected
or endogenous proteins—need to be addressed before firm conclusions regarding the impact of nutrient status on the stability
of the mTOR–Raptor complex in mammalian cells can be drawn. However, the following reports appear to make the scaffolding
model for Raptor function more attractive. Schalm and Blenis have identified a five amino-acid TOR-signaling (TOS) motif in
4E-BP1 and p70S6K that is required for mTOR-dependent phosphorylation of both proteins following the addition of nutrients
to starved cells (25). Whereas the TOS motif appears necessary for the binding of p70S6K or 4E-BP1 to Raptor (26 –28) , mutation of the TOS motif abolishes mTOR-catalyzed phosphorylation of 4E-BP1 in vitro in the presence of Raptor, and eliminates
the Raptor-dependent stimulation of mTOR-catalyzed p70S6K phosphorylation (26 , 27). Thus, the inhibitory effect of TOS deletion or mutation on the phosphorylation of 4E-BP1 or p70S6K in vivo can be attributed
to the inability of these mutants to bind Raptor. These results indicate one possible mechanism by which the Raptor couples
mTOR to cellular substrates.
New insights into bridging the phosphatidylinositol-3’ kinase (PI3K)–Akt signaling pathway and mTOR have emerged from recent
studies in both Drosophila melanogaster and mammalian cells involving the tumor suppressor proteins Tuberous Sclerosis Complex 1 and 2 (TSC1 and TSC2). The TSC1–TSC2
complex represses mTOR-dependent activation of p70S6K (29 –32) , and the putative GTPase-activating protein (GAP) domain of TSC2 inactivates the small GTPase Rheb (Ras homolog enriched
in brain) in vitro and in vivo (33 –36) (Figure 1⇑). TSC syndrome is an autosomal-dominant genetic disorder, characterized by mutations in either TSC1 or TSC2 that result in the widespread development of benign tumors termed hamartomas (37). In response to growth factor stimulation, the repression of mTOR signaling mediated by the TSC complex appears to be relieved
by Akt-mediated phosphorylation of TSC2, which requires the mitogen-induced activation of PI3K (30 , 31) (Figure 1⇑). Point mutations in the GAP domain of TSC2 disrupt its ability to regulate Rheb without affecting the ability of TSC2 to
form a complex with TSC1. Whether TSC1, TSC2, and/or Rheb bind to the mTOR complex containing Raptor–mLST8/Gβ L and modulate
the signaling function and kinase activity of mTOR remains an important but unanswered question. The further investigation
of Raptor, mLST8/Gβ L, and as-yet uncharacterized TOR-binding proteins will offer new targets for therapeutic invention in
human diseases, such as cancer and diabetes, in which mTOR signaling may be perturbed.
- © American Society for Pharmacology and Experimental Theraputics 2003
References
- ↵
Kunz, J., Henriquez, R., Schneider, U., Deuter Reinhard, M., Movva, N.R., and Hall, M.N. Target of rapamycin in yeast, TOR2,
is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 73 , 585–596 (1993).
- ↵
Cafferkey, R., Young, P.R., McLaughlin, M.M. et al. Dominant missense mutations in a novel yeast protein related to mammalian
phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity. Mol. Cell. Biol. 13 , 6012–6023 (1993). The article and the one above it identified of TOR proteins through genetic screening that revealed that mutations in TOR
genes rendered yeast resistant to rapamycin.
- ↵
Barbet, N.C., Schneider, U., Helliwell, S.B., Stansfield, I., Tuite, M.F., and Hall, M.N. TOR controls translation initiation
and early G1 progression in yeast. Mol. Biol. Cell. 7 , 25–42 (1996).
- ↵
Schmelzle, T. and Hall, M.N. TOR, a central controller of cell growth. Cell 103 , 253–262 (2000).
- ↵
Lin, T.A., Kong, X., Saltiel, A.R., Blackshear, P.J., and Lawrence, J.C., Jr. Control of PHAS-I by insulin in 3T3-L1 adipocytes.
Synthesis, degradation, and phosphorylation by a rapamycin-sensitive and mitogen-activated protein kinase-independent pathway.
J. Biol. Chem. 270 , 18531–18538 (1995).
- ↵
von Manteuffel, S.R., Gingras, A.C., Ming, X.F., Sonenberg, N., and Thomas, G. 4E-BP1 phosphorylation is mediated by the FRAP-p70S6K
pathway and is independent of mitogen-activated protein kinase. Proc. Natl. Acad. Sci. U.S.A. 93 , 4076–80 (1996). Important observations that concluded that TOR and p70S6K, rather than the expected mitogen-activated protein kinases (MAPKs)
were responsible activating 4E-BP1 and the initiation of translation.
- ↵
Chung, J., Kuo, C.J., Crabtree, G.R., and Blenis, J. Rapamycin-FKBP specifically blocks growth-dependent activation of and
signaling by the 70-kd S6 protein kinases. Cell 69 , 1227–1236 (1992).
- ↵
Price, D.J., Grove, J.R., Calvo, V., Avruch, J., and Bierer, B.E. Rapamycin-induced inhibition of the 70-kilodalton S6 protein
kinase. Science 257 , 973–977 (1992).
- ↵
Brown, E.J., Beal, P.A., Keith, C.T., Chen, J., Shin, T.B., and Schreiber, S.L. Control of p70 s6 kinase by kinase activity
of FRAP in vivo. Nature 377 , 441–446 (1995).
- ↵
Hara, K., Yonezawa, K., Kozlowski, M.T., Sugimoto, T., Andrabi, K., Weng, Q.P., Kasuga, M., Nishimoto, I., and Avruch, J.
Regulation of eIF-4E BP1 phosphorylation by mTOR. J. Biol. Chem. 272 , 26457–26463 (1997).
- ↵
Brunn, G.J., Hudson, C.C., Sekulic, A., Williams, J.M., Hosoi, H., Houghton, P.J., Lawrence, J.C., Jr., and Abraham, R.T.
Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science 277 , 99–101 (1997) The article and the two above it demonstrate that mTOR is an upstream regulator of p70S6K and 4E-BP1.
-
Burnett, P.E., Barrow, R.K., Cohen, N.A., Snyder, S.H., and Sabatini, D.M. RAFT1 phosphorylation of the translational regulators
p70 S6 kinase and 4E-BP1. Proc. Natl. Acad. Sci. U.S.A. 95 , 1432–1437 (1998).
- ↵
Isotani, S., Hara, K., Tokunaga, C., Inoue, H., Avruch, J., and Yonezawa, K. Immunopurified mammalian target of rapamycin
phosphorylates and activates p70 S6 kinase alpha in vitro. J. Biol. Chem. 274 , 34493–34498 (1999).
- ↵
Hara, K., Yonezawa, K., Weng, Q.P., Kozlowski, M.T., Belham, C., and Avruch, J. Amino acid sufficiency and mTOR regulate p70
S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 273 , 14484–14494 (1998). Results showed that sufficient concentrations of amino acids in the growth medium regulated the initiation peptide chain formation
during translation. Removing amino acids from the nutrient medium led to rapid deactivation of p70S6K and dephosphorylation
of 4E-BP1.
-
Wang, X., Campbell, L.E., Miller, C.M., and Proud, C.G. Amino acid availability regulates p70 S6 kinase and multiple translation
factors. Biochem. J. 334 , 261–267 (1998).
-
Patti, M.E., Brambilla, E., Luzi, L., Landaker, E.J., and Kahn, C.R. Bidirectional modulation of insulin action by amino acids.
J. Clin. Invest. 101 , 1519–1529 (1998).
-
Fox, H.L., Kimball, S.R., Jefferson, L.S., and Lynch, C.J. Amino acids stimulate phosphorylation of p70S6k and organization
of rat adipocytes into multicellular clusters. Am. J. Physiol. 274 , C206–C213 (1998).
-
Shigemitsu, K., Tsujishita, Y., Hara, K., Nanahoshi, M., Avruch, J., and Yonezawa, K. Regulation of translational effectors
by amino acid and mammalian target of rapamycin signaling pathways. Possible involvement of autophagy in cultured hepatoma
cells. J. Biol. Chem. 274 , 1058–1065 (1999).
- ↵
Xu, G., Marshall, C.A., Lin, T.A., Kwon, G., Munivenkatappa, R.B., Hill, J.R., Lawrence, J.C., Jr., and McDaniel, M.L. Insulin
mediates glucose-stimulated phosphorylation of PHAS-I by pancreatic beta cells. An insulin-receptor mechanism for autoregulation
of protein synthesis by translation. J. Biol. Chem. 273 , 4485–4491 (1998).
- ↵
Kim, D.H., Sarbassov, D.D., Ali, S.M., King, J.E., Latek, R.R., Erdjument-Bromage, H., Tempst, P., and Sabatini, D.M. mTOR
interacts with Raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110 , 163–175 (2002).
- ↵
Hara, K., Maruki, Y., Long, X., Yoshino, K., Oshiro, N., Hidayat, S., Tokunaga, C., Avruch, J., and Yonezawa, K. Raptor, a
binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110 , 177–189 (2002). Describes the characterization of the Raptor protein that is thought to act as a scaffolding protein that facilitates the
ability of mTOR to phosphorylate its substrates.
- ↵
Loewith, R., Jacinto, E., Wullschleger, S., Lorberg, A., Crespo, J., Bonenfant, D., Oppliger, W., Jenoe, P., and Hall, M.
Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10 , 457–468 (2002).
- ↵
Kim, D.H., Sarbassov, D.D., Ali, S.M., Latek, R.R., Guntur, K.V., Erdjument-Bromage, H., Tempst, P., and Sabatini, D.M. Gβ
L, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between Raptor
and mTOR. Mol. Cell 11 , 895–904 (2003). Article that posits GβL in a switch mechanism that regulates the interaction between mTOR and Raptor.
- ↵
Rodgers, B.D., Levine, M.A., Bernier, M., and Montrose-Rafizadeh, C. Insulin regulation of a novel WD-40 repeat protein in
adipocytes. J. Endocrinol. 168 , 325–332 (2001).
- ↵
Schalm, S.S. and Blenis, J. Identification of a conserved motif required for mTOR signaling. Curr. Biol. 12 , 632–639 (2002).
- ↵
Nojima, H., Tokunaga, C., Eguchi, S. et al. The mammalian target of rapamycin (mTOR) partner, Raptor, binds the mTOR substrates
p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J. Biol. Chem. 278 , 15461–15464 (2003).
- ↵
Schalm, S.S., Fingar, D.C., Sabatini, D.M., and Blenis, J. TOS motif-mediated Raptor binding regulates 4E-BP1 multisite phosphorylation
and function. Curr. Biol. 13 , 797–806 (2003).
- ↵
Choi, K.M., McMahon, L.P., and Lawrence, J.J. Two motifs in the translational repressor PHAS-I required for efficient phosphorylation
by mammalian target of rapamycin and for recognition by Raptor. J. Biol. Chem. 278 , 19667–19673 (2003).
- ↵
Manning, B., Tee, A., Logsdon, M., Blenis, J., and Cantley, L. Identification of the tuberous sclerosis complex-2 tumor suppressor
gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol. Cell 10 , 151–162 (2002).
- ↵
Inoki, K., Li, Y., Zhu, T., Wu, J., and Guan, K. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling.
Nat. Cell Biol. 4 , 648–657 (2002).
- ↵
Potter, C., Pedraza, L., and Xu, T. Akt regulates growth by directly phosphorylating Tsc2. Nat. Cell Biol. 4 , 658–665 (2002). This article and the one above it demonstrate that the PI3K pathway can regulate TSC-mediated activation of mTOR.
- ↵
Gao, X., Zhang, Y., Arrazola, P., Hino, O., Kobayashi, T., Yeung, R.S., Ru, B., and Pan, D. Tsc tumor suppressor proteins
antagonize amino-acid-TOR signalling. Nat. Cell Biol. 4 , 699–704 (2002).
- ↵
Stocker, H., Radimerski, T., Schindelholz, B., Wittwer, F., Belawat, P., Daram, P., Breuer, S., Thomas, G., and Hafen, E.
Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat. Cell Biol. 5 , 559–566 (2003).
-
Saucedo, L.J., Gao, X., Chiarelli, D.A., Li L, P.D., and Edgar, B.A. Rheb promotes cell growth as a component of the insulin/TOR
signalling network. Nat. Cell Biol. 5 , 566–571 (2003).
-
Zhang, Y., Gao, X., Saucedo, L.J., Ru, B., Edgar, B.A., and Pan, D. Rheb is a direct target of the tuberous sclerosis tumor
suppressor proteins. Nat. Cell Biol. 5 , 578–581 (2003).
- ↵
Garami, A., Zwartkruis, F.Z.T., Nobukuni, T. et al. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is
inhibited by TSC1 and 2. Mol. Cell. Plublished online June 2, 2003. 10.1016/S109727650300220X
- ↵
Sparagana, S.P. and Roach, E.S. Tuberous sclerosis complex. Curr. Opin. Neurol. 13 , 115–119 (2000). A review of the genetic mutations that lead to Tuberous Sclerosis Complex and the syndrome’s effects on various organs.
Kazuyoshi Yonezawa, MD, PhD, is a Professor at the Biosignal Research Center, Kobe University. Address correspondence to KY. E-mail yonezawa{at}kobe-u.ac.jp; fax +81-78-803-5970.





