Sites of interest on the World Wide Web—edited by Rick Neubig and David Roman
Resource for Pathways: Genes and Graphics
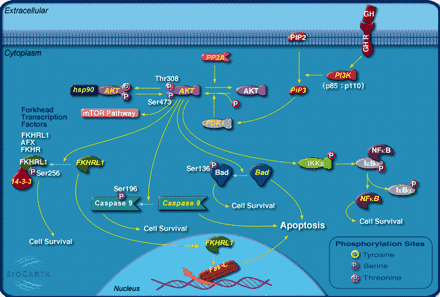
Located at http://www.biocarta.com/genes/index.asp and provided by BioCarta, this resource provides a very large number of graphical pathways of cellular signaling events. Ranging from Acetylation and Deacetylation of RelA in the Nucleus to Toll-like Receptor and WNT Signaling Pathways, this collection of clearly represented pathways—with great graphics—is sure to be a useful resource for teachers and researchers alike. Perhaps even more useful are the links to downloadable Macromedia Freehand and Adobe Illustrator format templates for the creation of a graphic of your favorite cell signaling pathway
Another site, the Kyoto Encyclopedia of Genes and Genomes (http://www.genome.ad.jp/kegg/), is home to data relating to thousands of genes, gene clusters, signaling pathways, chemical compounds, as well as knockout information. This Web site includes a huge list of pathway maps, grouped by organism, as well as software and development tools for microarray data analysis and compound drawing tools. The site is divided into easy-to-navigate search fields and lists of pathway links, and once you delve into your favorite area, the amount of information available is impressive. This Web site should be of use to those seeking more than just nice pathway pictures, for the genetic information background provides plenty of substance.
Cytochrome P450 Resource Pages
An article in this month’s Molecular Interventions, by Kharash et al., focuses on the use of drug probes for assessing CYP3A metabolic activity. A Directory of P450-Containing Systems (http://www.icgeb.trieste.it/~p450srv/), established by Kirill Degtiarenko, provides hyperlinked entries for GenBank and SwissProt accession numbers. Another repository of P450 information is maintained by Dr. David Nelson at the University of Tennessee Health Sciences Center in Memphis. The Cytochrome P450 Homepage (http://drnelson.utmem.edu/CytochromeP450.html) contains information and links to all types of P450 information, including databases, nomenclature, 3D P450 structures, links to P450 talks and meetings, and some interesting P450 statistics as well. This wellorganized and frequently updated resource (which has been highlighted previously in Net Results) provides a valuable collection of data and tools for exploring the large P450 family of enzymes.
Web-Based Molecular Biology Tool
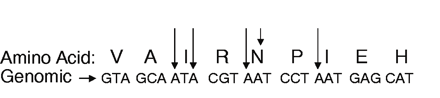
VectorDesigner is a Java-based Web tool that allows researchers to design, build, and view recombinant DNA molecules using virtual “cut-and-drop,” PCR, Gateway and TOPO cloning techniques. This graphics-based program from Invitrogen (https://vectordesigner.invitrogen.com) is available following free registration. The program allows for imported sequences, sequence annotation, restriction enzyme site identification, as well as primer designing capabilities. Registration also allows users to save sequences (primers, vectors, etc.) in their personal online database for later retrieval and manipulation. Those familiar with the VectorNTI software will feel right at home with the Web-based interface. As an added benefit, sequences and data files can be easily exchanged with other Web users, or users of the commercial Vector NTI software. Give this site a try, and you will be amazed at the quality of tools that are freely available for those tricky cloning projects.
- © American Society for Pharmacology and Experimental Theraputics 2005