Regulation of Receptors and Transporters by Ubiquitination: New Insights into Surprisingly Similar Mechanisms
Abstract
The function of many receptors and transport proteins that reside at the surface of the cell is regulated by endocytosis and postendocytic trafficking. Modification of receptors and transporters by ubiquitin conjugation has recently emerged as the major regulatory mechanism of internalization and intracellular sorting of these membrane proteins. This review will describe recent advances in elucidating the mechanisms of ubiquitination of mammalian receptors and transporters using two examples: the receptor for epidermal growth factor and the dopamine transporter. How ubiquitination controls the endocytosis and turnover of these proteins will be also discussed.
Introduction
Many integral membrane proteins, such as receptors, channels, ATPases, and transporters function at the plasma membrane where they interact with their ligands and substrates. Regulated expression of these membrane proteins at the cell surface is critical for their normal function. Transmembrane proteins are cotranslationally translocated into the endoplasmic reticulum (ER) and posttranslationally modified in the ER–Golgi membrane system before being transported to their sites of residence in the cell. Surface expression of membrane proteins is closely regulated by the quality control mechanisms in the ER and Golgi apparatus. Rapid and dynamic regulation of protein amounts at the plasma membrane, however, is achieved by means of selective endocytosis and recycling of these proteins. Many receptors and transport proteins rapidly internalize into endosomes in a constitutive or stimuli-dependent manner. Subsequently, the internalized proteins (i.e., cargo) are either recycled back to the plasma membrane or sorted to lysosomes for degradation. The mechanisms of endocytosis and postendocytic trafficking of membrane proteins have been extensively studied over the last thirty years; however, molecular details of many steps of these processes remain poorly understood.
Posttranslational modification of transmembrane proteins by the covalent attachment of ubiquitin has recently emerged as the major regulatory mechanism of endocytic trafficking of these proteins. Many of the original observations of ubiquitination of the endocytic cargo and regulation of endocytosis by ubiquitination were made in yeast (Saccharomyces cerevisiae), where various mutant strains deficient in ubiquitination and endocytosis have been generated (1, 2). These findings in yeast prompted a stream of studies in mammalian cells, which demonstrated that ubiquitination occurs in a variety of transmembrane proteins. Among mammalian ubiquitinated cargo are the receptor tyrosine kinases (RTKs); Notch and various cytokine and interferon receptors; various channels and transporters; G protein–coupled receptors (GPCR); and other types of transmembrane proteins. Several recent excellent reviews comprehensively describe the role of ubiquitination in the regulation of endocytic trafficking (3, 4); hence, we will focus on recent advances in understanding the mechanisms and functional roles of ubiquitination of two classes of pharmacologically important molecules: 1) RTKs, and 2) plasma membrane solute transporters. The first class was extensively utilized as the model system for elucidating the basic mechanisms and the role of ubiquitination in the regulation of endocytosis and endosomal sorting. The second class, transport proteins such as neurotransmitter transporters, represent an example of recently characterized ubiquitinated proteins for which the understanding of the function of ubiquitination has been rapidly advancing owing to the application of novel experimental methodologies.
Modification of Proteins by Ubiquitin
Ubiquitination (termed elsewhere as ubiquitylation or ubiquitinylation) is a posttranslational modification that mediates the covalent conjugation of ubiquitin to protein substrates. Ubiquitin is a highly conserved protein of seventy-six amino acids expressed in all eukaryotic cells. It contains a diglycine motif at the C-terminal end, in which the last residue, Gly76, can be conjugated to the ε-amino group of a lysine residue in a target protein (Figure 1⇓). The functional role of ubiquitination was originally thought to target cytosolic proteins for degradation by the 26S proteasome (5). However, it is now known that ubiquitination regulates many other processes in the cell, including membrane trafficking, DNA repair, and transcription (6, 7).
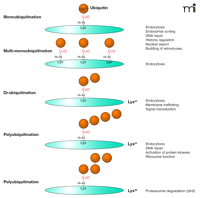
Types of ubiquitin conjugation. Gly76 of ubiquitin is covalently attached to the ε-amino group of lysines in the substrate. Substrates can be modified with a single ubiquitin molecule at a single (monoubiquitination) or multiple (multi-monoubiquitination) lysine residues. Further ubiquitin conjugation to the lysine residues of ubiquitin results in diubiquitin or polyubiquitination. Shown are the most frequently detected ubiquitin chains linked through Lys63 or Lys48 of ubiquitin. Lys48- or Lys63-linked chains have closed and extended conformation, respectively, resulting in different mechanisms of recognition by ubiquitin binding domains (UBDs).
The mechanism of ubiquitination involves the sequential action of several enzymes. In the initial step, the E1 ubiquitin-activating enzyme forms a thioester bond between its catalytic cysteine and the carboxyl group of Gly76 of ubiquitin in an ATP-dependent manner. The ubiquitin molecule is then transferred to an E2 ubiquitin-conjugating enzyme, which also forms a thioester bond between its cysteine and ubiquitin. Finally, ubiquitin is transferred to a lysine residue of the substrate with the help of an E3 ubiquitin ligase. There are only one or perhaps two E1 enzymes, and fewer than sixty E2 enzymes and more than 400 E3 ligases in the human genome (8). Such an abundance of E3 ligases allows these proteins to provide the specificity of target recognition in the ubiquitin-ligase system (9). The isopeptidases responsible for removal of ubiquitin from the substrate are called deubiquitination enzymes (DUBs) (10). DUBs may prove to be as important as the ubiquitin ligases are for regulating ubiquitin-mediated effects; however, functions of DUB and their substrate specificity are poorly studied as compared to that of the ligase enzymes.
Attachment of a single ubiquitin moiety to a single lysine on a substrate results in monoubiquitination (Figure 1⇑). Monoubiquitination of several lysine residues of a protein results in multi-monoubiquitination. Additional ubiquitin molecules can be attached to the lysine residues in ubiquitin itself, resulting in the formation of diubiquitin and polyubiquitin chains on a single lysine of the substrate. Although ubiquitin contains seven lysine residues, all capable of conjugating ubiquitin, Lys48- and Lys63-linked chains are the most abundant. The majority of published studies suggest that Lys48-linked chains serve as the recognition signal by the proteasome and target proteins for proteasomal degradation (6). In contrast, Lys63-linked ubiquitin chains do not target proteins to proteasome but play important roles in interactions with protein machineries involved in endocytic trafficking, inflammatory response, protein translation, and DNA repair (6). Similarly, it is widely accepted that monoubiquitination does not target proteins to the proteasome but serves as a molecular recognition signal in membrane trafficking, regulation of endocytic machinery, and possibly other cellular processes (4).
All of these functions of ubiquitin are accomplished through specific interactions of the ubiquitin moiety with the ubiquitin binding domains (UBDs) found in many proteins (11). All of the helical UBDs interact with hydrophobic Ile44 in ubiquitin, although there are several types of UBD that have different modes of recognition of mono- and polyubiquitin (12). Recent studies demonstrated that Lys48-linked diubiquitin has a closed conformation, whereas Lys63-linked diubiquitin has an extended conformation, thus implying their selective recognition by different types of UBDs (13–15).
Stimuli-induced Ubiquitination of Receptors
The platelet-derived growth factor receptor (PDGFR) and the epidermal growth factor receptor (EGFR) were the first mammalian receptors found to be ubiquitinated (16, 17). Since these original reports, several other RTKs such as ErbB3 and ErbB4 (18), c-kit (19), TrkA (20, 21), c-Met/hepatocyte-growth-factor (HGF) (22), vascular endothelial growth factor (23), colony-stimulating factor (CSF1) (24), insulin-like growth factor 1 (IGF-1) (25), and other receptors have been observed to be ubiquitinated in a ligand-dependent manner. Thus, the regulatory function of ubiquitination has been proposed for at least six subfamilies of RTKs.
Elucidation of the signaling pathways that include the EGFR or PDGFR led to the discovery that the Cbl family of proteins––present in humans as three isoforms, c-Cbl, Cbl-b, and Cbl-c (26)––are E3 ubiquitin ligases. All three Cbls have an N-terminal tyrosine kinase binding (TKB) domain connected (with a linker segment) to a RING finger domain. c-Cbl and Cbl-b each have an extended C-terminal tail containing proline-rich motifs and an UBD. Cbl-c lacks most of the C-terminal region downstream from the RING domain. The TKB domain directly binds to the specific phosphotyrosine-containing motifs in RTK and other tyrosine kinases. The RING domain of the E3 ubiquitin ligase recruits an E2 and positions it so that the ubiquitin moiety can be transferred from E2 to the substrate. The proline-rich motifs in the Cbl C terminus mediate the interaction of Cbl with Src Homology 3 (SH3) domain–containing proteins.
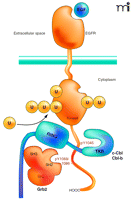
The putative mechanism of Cbl interaction with an RTK was proposed mainly on the basis of studies of the EGF and HGF/c-Met receptors (22, 27–29). Activation of the receptor kinase upon ligand binding leads to the phosphorylation of tyrosine residues in the C terminus of the receptor that serve as binding sites for the TKB domain of Cbl and the SH2 domain of Grb2 adaptor protein (Figure 2⇓). Grb2 is constitutively associated with the proline-containing motifs of Cbl via its SH3 domain, and is recruited to activated receptors as a complex with Cbl. Activation of receptors leads to tyrosine phosphorylation of Cbl either by the receptor kinase directly or through the kinase activity of c-Src (26, 30). Both direct and Grb2-mediated interactions of Cbl with the EGFR or c-Met are necessary for the full ubiquitination of these receptors.
Interactions of the EGF receptor leading to receptor ubiquitination. EGF binding activates the receptor tyrosine kinase and results in phosphorylation of Tyr1045, Tyr1068, and Tyr1086 in the C terminus of the receptor. The SH3 domains of Grb2 are associated with the C terminus of c-Cbl or Cbl-b. A Grb2-Cbl complex binds to the receptor by means of the interaction of the SH2 domain of Grb2 with phosphorylated Tyr1068 or phosphorylated Tyr1086 (pY1068/86) and the tyrosine kinase binding (TKB) domain of c-Cbl or Cbl-b interacts with the phosphotyrosine 1045 (pY1045) of the receptor. Recruitment of E2 (not shown) to the RING domain of Cbl results in covalent attachment of monoubiquitin and polyubiquitin chains to the kinase domain of the receptor.
The development of highly sensitive and quantitative methods of mass-spectrometry have allowed the characterization of ubiquitin modifications on the EGFR. Surprisingly, mass-spectrometry analysis revealed that the major sites of EGFR ubiquitination are located within the conserved kinase domain of the receptor (31). Additionally, in the absence of the major conjugation sites, other lysines became ubiquitinated, suggesting that EGFR ubiquitination sites are highly redundant. Importantly, quantitative mass-spectrometry analysis showed that EGFRs are both mono- and polyubiquitinated and that the most abundant type of polyubiquitination is the Lys63-linked chains (31). Similar analysis of other ubiquitinated RTKs has not been performed.
What is the functional role of RTK ubiquitination? Growth factor activation of many types of RTKs results in the accelerated turnover and decreased amounts of the receptor protein (32). Receptor “down regulation” serves as the major negative feedback regulatory mechanism of receptor signaling, and receptor ubiquitination appears responsible for much of this decreased expression. Indirect evidence is based on the observations that mouse knock-out of c-Cbl or overexpression of Cbl mutants, including naturally-occurring mutants of c-Cbl, 70Z-Cbl, or v-Cbl, leads to reduced ubiquitination and to slow turnover of EGFRs (27, 33, 34); and vice versa, overexpression of Cbl leads to elevated ubiquitination and accelerated turnover of the EGFR protein (34). Similar observations were made in experiments with several other RTKs (19, 22–24, 35).
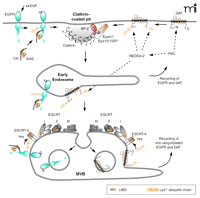
The two steps of trafficking that determine the rate of RTK turnover are internalization and postendocytic sorting to lysosomes (Figure 3⇓). The mechanisms of internalization and the role of receptor ubiquitination in this process are not well understood. Nonetheless, the binding of Cbl to the receptor via the adaptor protein Grb2 plus an intact Cbl RING domain are necessary for EGFR internalization through clathrin-coated pits, a physiological pathway of EGFR endocytosis (28, 29, 36). However, mutation of the major ubiquitination sites in the EGFR had no effect on its internalization (31). These results imply that either a minimal extent of ubiquitination (minor sites?) is sufficient for internalization, or the Cbl RING domain mediates internalization via the regulation of a process other than receptor ubiquitination.
Hypothetic model of endocytosis and endosomal sorting of the EGF receptor and DAT. (Top left) Binding of epidermal growth factor (EGF) to the EGF receptor (EGFR) results in recruitment of the Grb2-Cbl complex and receptor ubiquitination. EGFR is internalized via clathrin-coated pits with participation of Grb2 and Cbl by an unknown mechanism. After fusion of clathrin-coated vesicles with early endosomes (middle), EGFR can either recycle directly back to the plasma membrane or remain in the maturing endosome that acquires ESCRT complexes. Ubiquitinated receptors bind to the UBD of the ESCRT-0 complex, and eventually become trapped in the intralumenal vesicles of MVB (bottom). Non-ubiquitinated receptors can recycle back to the cell surface through the tubular extensions of MVB. (Top right) Activation of protein kinase C (PKC) results in NEDD4-2–mediated ubiquitination of the dopamine transporter (DAT). PKC activation can facilitate NEDD4-2–mediated ubiquitination of DAT either by phosphorylating DAT or DAT-interacting proteins, or by activating NEDD4-2. Ubiquitinated DAT is recruited into clathrin-coated pits by means of interaction with the UBD-containing proteins, such as Eps15/Eps15R and epsin, bound to AP-2 and clathrin in coated pits. PKC-dependent sorting of DAT in early endosomes (middle) and MVB (bottom) is hypothesized to occur by the mechanism similar to that of the EGFR. See text for details.
After internalization into early endosomes, receptors are either recycled back to the plasma membrane or sorted to the lysosome degradative pathway concomitantly with the maturation of endosomes. Because weakly ubiquitinated mutants of the EGFR are not efficiently degraded, proper ubiquitination might be critical for the lysosomal targeting of the receptor (27, 31). After EGF-induced endocytosis, EGF and EGFRs accumulate in the intralumenal vesicles of multi-vesicular endosomes or bodies (MVBs) that are mostly located in the perinuclear area of the cell (Figure 3⇑) (37, 38). EGFRs that are incorporated into intralumenal vesicles cannot recycle. MVBs have tubular membrane extensions that are thought to be responsible for recycling of receptors not recruited into internal vesicles (39).
The molecular mechanisms involved in sorting ubiquitinated EGFRs in MVBs have been, to a large extent, extrapolated from the genetic analysis of MVB sorting in yeast. This analysis defined molecular complexes, termed endosomal sorting complex required for transport (ESCRTs), which mediate the sorting process in MVBs (40). The ubiquitinated EGFR in endosomes is thought to interact with the UBD of the Hepatocyte growth factor receptor phosphorylation substrate (Hrs) that is associated with another UBD-containing protein, either STAM1 or STAM2, resulting in the ESCRT-0 complex (41). It is hypothesized that the ubiquitinated EGFR can be moved, in sequence, to ESCRTI, II, and III complexes, although binding of the EGFR to these complexes has not been demonstrated and the exact mechanism of this process is not understood (42). Components of ESCRTI and III complexes, TSG101 and hVps24, respectively, are necessary for EGFR degradation (43–45), whereas the requirement of the ESCRTII complex has not been confirmed in RNA interference experiments (45). Invagination of the limiting membrane of MVB is believed to be facilitated by the oligomerization of charged multivesicular body proteins (CHMPs, a component of ESCRTIII) and the formation of a lattice-like structure (40). Membrane invagination results in the formation of internal vesicles containing EGFRs. Based on studies in yeast, it has been proposed that cargo is deubiquitinated prior to its being trapped into internal vesicles, which may serve to prevent the degradation of ubiquitin (42); however, EGFR deubiquitination during MVB sorting has not been demonstrated.
MVBs fuse with primary lysosomal vesicles that contain proteolytic enzymes, which leads to the formation of mature lysosomes and rapid proteolysis of intralumenal components of MVBs, including EGFR (38). Degradation of EGF and the EGFR is completely blocked by lysosomal inhibitors, suggesting that it occurs in lysosomes (46, 47). Although the use of proteasomal inhibitors can also reduce EGFR degradation (48), these inhibitors may affect the activity of lysosomal enzymes, turnover of ESCRT proteins, or reduce the ubiquitin pool in the cell. Therefore, the effects of proteasomal inhibitors on EGFR degradation are likely indirect.
Whereas several subfamilies of RTK are regulated by Cbl-mediated ubiquitination, some RTKs appear to utilize other E3 ubiquitin ligases. For example, ErbB3 and ErbB4, which share a high degree of sequence similarity with EGFR, are constitutively ubiquitinated by the RING domain–containing ligase termed neuregulin receptor degradation protein-1 (Nrdp1) (18). Ligand (neuregulin) binding to these receptors results in elevated expression of Nrdp1 and, therefore, increased ubiquitination of ErbB3 and ErbB4, leading to decreased amounts of ErbB3 and ErbB4 (18). The molecular determinants for the interaction with Nrdp1 in ErbB receptors are unknown.
Ubiquitination of TrkA, a receptor for nerve growth factor, has been recently reported and implicated in the regulation of TrkA endocytosis (20, 21). TrkA participates in the development of the central nervous system and survival signaling in adult neurons. Internalization of TrkA in distal axonal processes is important for their retrograde signaling to neuronal soma (49). There is disagreement as to which E3 ubiquitin ligase is involved. One study proposed that the TRAF6 ubiquitin ligase is responsible for TrkA ubiquitination, mediated by the interaction of TrkA with the p75NTR co-receptor (20). It is noteworthy that, similar to the EGFR, the TrkA was proposed to be polyubiqutinated by Lys63-linked chains, which was shown to be critical for internalization (20). In contrast, another study claimed that TrkA is ubiquitinated by the E3 termed neuronal precursor cell expressed developmentally downregulated (NEDD4-2), which contains a homologous to E6-AP C-terminal (HECT) domain (21). Although the data regarding the TrkA-specific ubiquitin ligase are conflicting, both studies suggest that ubiquitination mediates endocytosis of TrkA and therefore affects signal transduction by this RTK. The methods of measuring the rate parameters of TrkA trafficking in these studies, however, did not have sufficient resolution to determine whether inhibition of TrkA ubiquitination resulted in decreased internalization or increased recycling (owing to the reduced lysosomal targeting), or both.
Mammalian GPCR represent another class of pharmacologically important receptors that have also been found to be ubiquitinated upon agonist stimulation (50). In the cases of the β2-adrenergic receptor and the chemokine receptor CXCR4, it was possible to directly test the role of receptor ubiquitination because all cytoplasmic lysines could be mutated without affecting the biological activity of the receptors (51, 52). These experiments showed that receptor ubiquitination was not necessary for internalization but was critical for endosomal sorting to the lysosomal degradative pathway. The identity of the E3 ubiquitin ligase responsible for ubiquitination of the β2-adrenergic receptor is not known. A HECT domain–containing E3 ligase, AIP4, has been reported to regulate trafficking of CXCR4, whereas Cbl might ubiquitinate protease-activated receptor 2 (PAR2) (53, 54). Agonist binding also stimulates ubiquitination of GPCR-associated proteins (e.g., non-visual β-arrestins 1 and 2) that function as adaptors linking activated GPCR to clathrin-coated pits (51). In contrast to receptor ubiquitination, ubiquitination of β-arrestins has been proposed to be necessary for receptor internalization (51). The mechanisms of lysosomal targeting of ubiquitinated GPCR are thought to be similar to that described above for the EGFR sorting in MVBs, although the data describing the morphology and biochemistry of GPCR sorting in MVBs are rather limited. Importantly, there are examples of GPCR, such as δ-opioid and PAR1 receptors, that are not ubiquitinated in an agonist-dependent manner but efficiently degraded in lysosomes (55, 56). The mechanism of lysosomal targeting of these receptors is unclear.
Regulation of Plasma Membrane Transporters by Ubiquitination
Membrane transport proteins have essential roles in fundamental cellular processes such as the maintenance of ion homeostasis, uptake of nutrients, neurotransmission, signal transduction, elimination of toxic compounds, cell excitability, and intercellular communication. Thus, regulation of transporter endocytosis and post-endocytic trafficking has a significant impact on cell physiology. An ATP-binding cassette (ABC) transporter, Ste6, required for secretion of the mating pheromone α-factor in yeast, was the first identified plasma membrane transport protein whose surface expression was regulated by ubiquitination (1). Subsequently, endocytosis of several plasma membrane transporters in yeast is now known to be regulated by ubiquitination. The role of ubiquitination in internalization and post-endocytic targeting to the vacuole has been best characterized for the general amino acid (Gap1) and uracil (Fur4) permeases (57–59). Ubiquitination, internalization, and vacuole-dependent degradation of these transporters require the HECT domain–containing E3 ubiquitin ligase Rsp5, a yeast homolog of mammalian NEDD4-2 [reviewed in (60)]. These permeases/transporters are modified with Lys63-linked polyubiquitin chains. Blocking the formation of Lys63-linked chains by overexpression of a ubiquitin mutant incapable of forming these chains partially inhibited internalization and degradation of Gap1 and Fur4 (58, 61).
The list of mammalian transport proteins that are ubiquitinated continues to grow (Table 1⇓). The common theme seems to be that ubiquitination controls the function of various channels, ATPases, and secondary transporters by regulating internalization and/or lysosomal degradation of these proteins. The amiloride-sensitive epithelial sodium channel (ENaC) was the first mammalian transport protein found to be ubiquitinated (62). The mechanisms of ENaC ubiquitination and regulation of its endocytosis by ubiquitination have been recently reviewed (4, 63). Hence, we focus on the most recent studies of another type of membrane transport protein, the dopamine (DA) transporter (DAT). To our knowledge, DAT is the only transporter for which several novel technologies, such as RNA interference library screening and quantitative mass spectrometry, have been used to: 1) demonstrate ubiquitination of the transporter; 2) map major ubiquitination sites; 3) identify its E3 ligase; and 4) define the function of ubiquitination in the endocytosis of this transporter.
Mammalian Plasma Membrane Transport Proteins that Undergo Ubiquitination
DAT is expressed in dopaminergic neurons in the brain, where it functions to terminate DA neurotransmission via the reuptake of released DA into dopaminergic neurons. DAT is implicated in the development of neurodegenerative disorders and drug abuse (64). DAT belongs to the SLC6 family of Na+-Cl−-dependent co-transporters and has twelve transmembrane domains and intracellular N and C termini (65). Mass spectrometry analysis of purified human DAT revealed that DAT is constitutively ubiquitinated and that activation of protein kinase C (PKC) substantially increases DAT ubiquitination. Furthermore, mass spectrometry also revealed the presence of Lys63-linked polyubiqutin chains in DAT, and immunoblot analysis predicted that each DAT molecule is conjugated at any given time with a single short chain of two or three ubiquitins (66). Similarly, Lys63-linked polyubiquitination of the aquaporin-2 (AQP-2) water channel has been recently reported (67).
RNA interference analysis revealed that PKC-dependent DAT ubiquitination requires NEDD4-2 (68), which has been implicated in the ubiquitination of various transport proteins, including ENaC channels (69) (Table 1⇑). The NEDD4 family of proteins is characterized by the presence of a C-terminal HECT domain, N-terminal C2 domain that binds phospholipids in a Ca2+-dependent manner, and two to four WW domains that bind to the PPxY (PY) motif (x is any amino acid) (4). PY motifs are found in the C-terminal tails of various transmembrane proteins. For instance, the carboxyl-terminal regions of each subunit (α, β, γ) of ENaC contain PY motifs, which interact with the WW domains of NEDD4-2 (70). This interaction is necessary for ubiquitination of the α and γ subunits (62). However, a number of transporters that are regulated by NEDD4-2, including DAT, lack the PY motif. It is possible that NEDD4-2 binds indirectly to these transporters, in a manner similar to that described for the IGF-1 receptor (71). Another possibility is that NEDD4-2 may regulate another E3 ligase that directly ubiquitinates transporters.
PKC-induced DAT ubiquitination takes place initially at the plasma membrane and continues after endocytosis. Mass-spectrometry analysis enabled mapping the major ubiquitination sites in the N and C termini of DAT (66). Mutagenesis of these lysines revealed that a cluster of three N-terminal lysines are essential for PKC-dependent endocytosis of DAT (72). Moreover, RNA interference analysis showed that DAT is internalized via a clathrin-mediated pathway and that this internalization is dramatically inhibited by mutation of the ubiquitination sites (72, 73). Finally, this PKC-dependent internalization of DAT required NEDD4-2 and the adaptor proteins epsin, Eps15, and Eps15R that are located in clathrin coated pits and possess UBDs (Figure 3⇑) (68). Similarly, epsin and Eps15 have been recently shown to be involved in NEDD4-2 dependent internalization of ENaC (74).
The methods of measuring the rate parameters of endocytic trafficking of DAT and other transporters do not allow quantification of the internalization rates without the contribution of recycling. Therefore, the steps of endocytic trafficking of transporters that are regulated by ubiquitination are not always precisely defined. Whereas several sets of data suggest that activation of PKC results in accelerated internalization of DAT in a ubiquitin-dependent manner, it also leads to accelerated degradation of DAT in lysosomes (66, 75). Therefore, it is likely that the sorting of DAT to lysosomal degradation versus recycling pathway is also mediated by ubiquitination. As described above for the EGFR model (Figure 3⇑), this sorting probably involves trapping the transporters inside MVB. Co-localization of DAT with ESCRT proteins in endosomes (66, 76) and detection of DAT inside the MVB in neurons support this notion (77). The essential role of ubiquitination in sorting to lysosomes of other mammalian transport proteins, such as ENaC and AQP2, has also been proposed (62, 67, 78). Overall, more detailed structure-function and electron microscopy studies should be performed to characterize the role of NEDD4-2 and ubiquitination in the intracellular sorting of transporters. However, a striking similarity of the regulation of these processes among various receptor and transporter proteins is already quite evident (Figure 3⇑).
Conclusions and Outstanding Issues
During the last decade, ubiquitination has emerged as the major mode of regulation of the subcellular localization and turnover of membrane proteins, many of which are implicated in human disease and represent important therapeutic targets. The general consensus is that ubiquitination of the integral membrane proteins mediates the post-endocytic sorting of these proteins to lysosomes. In contrast, the role of ubiquitination in internalization of these proteins from the plasma membrane has been directly demonstrated only in few cases in mammalian cells. The notion that these regulatory functions of ubiquitination are mediated by mono-ubiquitination has been dominant in the literature. It is now clear, however, that Lys63-linked polyubiquitination is the common modification of many types of transmembrane proteins. Although further investigation is needed to examine the precise role of Lys63-linked chains in endocytic trafficking, it can be speculated that whereas monoubiquitin binds to most UBDs with the low affinity, the linear conformation of Lys63-linked chains allows multivalent interactions of the UBD-containing proteins with Lys63-polyubiqutinated cargo, thus increasing the avidity of the interaction.
Two model systems, the EGFR and DAT, discussed here as examples of ubiquitinated endocytic cargo, point out the fact that a large portion of the data describing these models have been obtained under experimental conditions that differ from the conditions in vivo. For instance, it remains to be tested whether stimulation of the cells with the physiological concentration of the EGFR ligand would result in EGFR ubiquitination, interaction with ESCRT complexes, and sorting in MVB (Figure 3⇑). Likewise, it is unclear whether DAT is ubiquitinated and regulated by PKC in dopaminergic neurons in a manner similar to that observed in non-neuronal expression systems. In this respect, it would be interesting to investigate if a psychostimulant like amphetamine that is known to activate PKC would trigger ubiquitination and ubiquitin-dependent trafficking of DAT in the intact brain and if such a ubiquitination contributes to the abuse potential of this compound. Future studies would also test whether ubiquitin-dependent trafficking is the general mechanism of the regulation of neurotransmitter and other solute transporters.
The ability to pharmacologically intervene with ubiquitination of the receptors and transporters would depend on the generation of specific inhibitors of E3 ubiquitin ligases or DUBs. In most cases the specific DUB responsible for deubiquitination of a substrate is unknown. Because E3 ligases display substantial specificity towards substrates or a family of substrates, such as Cbl towards an RTK family, an attractive pharmacological target could also be a protein that modulates an E3. Several putative modulators of Cbl have been recently identified (79). The activity of NEDD4-2 ligase is known to be negatively regulated by serum and glucocorticoid-regulated kinases (4). Development of therapeutics capable of manipulating the ubiquitination of receptors and transporters would require further elucidation of the molecular mechanisms of the specific interactions of these membrane proteins with ubiquitin ligases, DUBs, and other proteins regulating the cellular ubiquitination system.
Acknowledgments
We thank Melissa Adams and Arnau Vina for the help in preparation of the manuscript. The work of the authors is supported by National Cancer Institute, American Cancer Society and National Institute of Drug Abuse.
- © American Society for Pharmacology and Experimental Theraputics 2007
References

Alexander Sorkin, PhD, is a Professor of Pharmacology at the University of Colorado Health Sciences Center. His work focuses on the mechanisms of endocytosis of the epidermal growth factor receptor and the dopamine transporter, and the role of endocytic trafficking of these proteins in their functions. Address correspondence to AS. E-mail Alexander.sorkin{at}uchsc.edu; fax 303-724-3663.

Manuel Miranda, PhD, is a senior postdoctoral fellow in the Department of Pharmacology at the University of Colorado Health Sciences Center. His research throughout his career has focused on studying the structure-function and trafficking mechanisms of ion transporters. He has been recently appointed as an Assistant Professor at the University of Texas at El Paso (UTEP).