
- Institution: Stanford Univ Med Ctr Lane Med Lib/Periodical Dept/Rm L109
- Sign In as Member / Individual
Anti-HIV-1 Therapeutics: From FDA-approved Drugs to Hypothetical Future Targets
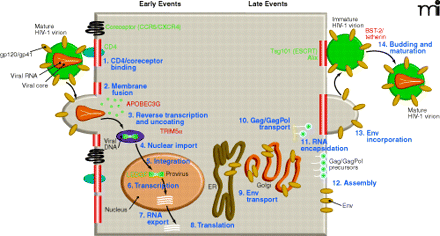
The HIV-1 replication cycle. Major steps of HIV-1 replication are numbered. The HIV-1 replication cycle begins with viral entry into the target cell. Entry occurs upon fusion of the viral lipid envelope with the host cell plasma membrane, a process mediated by a non-covalent complex of viral (Env) glyco-proteins gp120 and gp41. First, gp120 binds the cellular receptor CD4, and then interacts with the CCR5 or CXCR4 coreceptor. Coreceptor binding by gp120 triggers a series of conformational changes in both gp120 and gp41 that lead to membrane fusion. Following fusion, the viral core, composed of a capsid (CA) shell containing the dimeric single-stranded RNA genome in complex with the reverse transcriptase (RT) and integrase (IN) enzymes, is deposited into the cell. The core uncoats, and RT copies the RNA genome into a double-stranded DNA copy. This viral DNA is then transported into the cell nucleus where it is integrated into the host cell genome by the IN enzyme. Subsequent transcription and translation lead to production of the viral components that assemble into new particles. The assembly process, which occurs at the plasma membrane, is directed by the Gag polyprotein precursor. To promote the budding and release of newly assembled virus particles, HIV-1 hijacks cellular endosomal sorting machinery (referred to as the “ESCRT” complexes) that normally functions to promote the budding of vesicles into late endosomes to form multivesicular bodies. Following release, the viral protease (PR) cleaves the Gag polyprotein precursor into individual Gag domains, thereby triggering a maturation process that is absolutely required for particle infectivity. Host factors that play a positive role in HIV-1 replication are indicated in green; factors that can restrict HIV-1 replication are in red. Adapted and reprinted with permission from Elsevier (1), © 2004.


