|
 
 |
| ORIGINAL ARTICLE |
|
| Year : 2014 | Volume
: 9
| Issue : 4 | Page : 151-156 |
|
Microbiological studies of waste dumpsite in Abia State University Teaching Hospital, Aba
Eugene Chimezie Ndimele1, Uzochukwu Godswill Ekeleme2, Alloysius Chibuike Ogodo3, Nkechi Chinyere Nwachukwu4, Chidiebere James Nnadi4, Elijah Akachukwu Otutu4
1 Department of Primary Health Care, Faculty of Medicine and Health Sciences, Abia State University, P.M.B. 2000, Uturu, Abia State, Nigeria
2 Department of Biology and Microbiology, Faculty of Natural and Applied Sciences, Gregory University, P.M.B. 1012, Achara, Uturu, Abia State, Nigeria
3 Department of Microbiology, Faculty of Pure and Applied Sciences, Federal University, P.M.B. 1020, Wukari, Taraba State, Nigeria
4 Department of Microbiology, Faculty of Biological and Physical Sciences, Abia State University, P.M.B. 2000, Uturu, Abia State, Nigeria
| Date of Web Publication | 14-May-2015 |
Correspondence Address:
Dr. Eugene Chimezie Ndimele
Department of Primary Health Care, Faculty of Medicine and Health Sciences, Abia State University, P. M. B. 2000, Uturu, Abia State
Nigeria
 Source of Support: None, Conflict of Interest: None  | Check |
DOI: 10.4103/9783-1230.157058

Background: Hospital is a vital place for our life, health and well-being. However, the waste generated from hospital can be hazardous, toxic and even lethal because of their high potential for disease transmission. Objective: The work is aimed at doing microbiological studies of hospital waste dumpsite in Abia State University Teaching Hospital (ABSUTH), Aba. Materials and Methods: The study area was ABSUTH, Aba. Passive air sampling was performed using settle plates, wastewater samples from the drippings, soil sediment underlying solid waste and soil adjacent to the dumpsite were collected from a depth of 0.9-30 cm and was evaluated using the standard microbiological and molecular techniques. Result: The result of the total microbial count of air samples within the waste dumpsite and reception unit of ABSUTH, Aba revealed that the total viable count (TVC) was 182 ± 0.5 air within waste dumpsite (AW) and 70 ± 2.0 air within reception unit (AR), the total coliform count (TCC) was 17 ± 0.2 (AW) and 5 ± 1.0* (AR), the total staphylococcal count (TSC) was 8 ± 0.1 (AW) and 3 ± 0.5* (AR) while the total fungal count was 60 ± 0.4 (AW) and 42 ± 0.7 (AR), the variations in the results differ significantly at P < 0.05. The total microbial count of soil samples within the waste dumpsite showed that the TVC was 3.0 ± 1.7 × 10 8 cfu/g which was significantly different (P < 0.05) when compared with the control, 3.2 ± 0.8 × 10 6 cfu/g. The TCC was 1.6 ± 0.4 × 10 4 cfu/g and do not differ significantly (P > 0.05) with the control (1.0 ± 1.0 × 10 4 cfu/g), the TSC was 2.6 ± 0.7 × 10 2 cfu/g and do not differ significantly (P > 0.05) with the control (2.1 ± 0.1 × 10 2 cfu/g) while the total fungal counts (6.8 ± 0.3 × 10 7 cfu/g) showed a significant difference when compared with the control (5.3 ± 0.6 × 10 6 cfu/g). The total microbial count of dripping samples from the waste dumpsite of ABSUTH showed that the TVC, TCC, staphylococcal count and the fungal count were 6.9 ± 1.7 × 10 7 , 3.5 ± 1.0 × 10 3 , 1.8 ± 0.6 × 10 2 , and 7.4 ± 0.1 × 10 5 , respectively. The variations in microbial counts of the dripping hospital waste dumpsite samples differed significantly (P < 0.05) when compared with the controls which showed no growth. The most occurring microorganisms were Bacillus species, Staphylococcus aureus, Streptococcus sp. and Aspergillus fumigatus. The susceptibility profile of bacterial isolates revealed that ciprofloxacin and augmentin produced the highest percentage inhibition of 33 to 100% against all the bacterial isolates except Actinomycetes isreali which was resistant. Most of the isolates had no plasmid except for Escherichia coli which produced equal size of 23 kb of plasmid. Conclusion/Recommendation: The high microbial load densities suggests that the hospital wastes in the environment pose a major health and environmental threat. This study therefore calls for a proper regulatory system on disposal of hospital waste.
Keywords: Antibiotics and plasmid, hospital, microorganisms
How to cite this article:
Ndimele EC, Ekeleme UG, Ogodo AC, Nwachukwu NC, Nnadi CJ, Otutu EA. Microbiological studies of waste dumpsite in Abia State University Teaching Hospital, Aba. J Med Investig Pract 2014;9:151-6 |
How to cite this URL:
Ndimele EC, Ekeleme UG, Ogodo AC, Nwachukwu NC, Nnadi CJ, Otutu EA. Microbiological studies of waste dumpsite in Abia State University Teaching Hospital, Aba. J Med Investig Pract [serial online] 2014 [cited 2018 Aug 24];9:151-6. Available from: http://www.jomip.org/text.asp?2014/9/4/151/157058 |
| Introduction | |  |
Hospital is a vital place for our life, health and well-being. But the waste generated from hospital can be hazardous, toxic and even lethal because of their high potential for disease transmission. The hazardous and toxic parts of the waste from health care establishments comprising infectious, biomedical and radioactive material as well as sharps (hypodermic needles, knives, scalpels etc.) constitute a grave risk, if these are not properly treated or disposed of or if is allowed to get mixed with other municipal waste. Its propensity to encourage growth of various pathogen and vectors and its ability to contaminate other nonhazardous or nontoxic municipal waste jeopardizes the efforts undertaken for overall municipal waste management. The rag pickers and waste workers are often the worst affected, because unknowingly or unwittingly, they rummage through all kinds of poisonous material while trying to salvage items which they can sell for reuse. At the same time, this kind of illegal and unethical reuse can be extremely dangerous and even fatal. Diseases like cholera, plague, tuberculosis, hepatitis (especially hepatitis B virus), AIDS (HIV), diphtheria etc., in either epidemic or even endemic form, pose grave public health risks. Hospital solid waste (SW) can contain significantly high concentrations of pathogenic organisms. [1],[2] This effect is increased if there is inadequate handling of these wastes. Infectious waste may contain pathogens, which can infect people through a number of routes, such as punctures, abrasions, or lacerations of the skin, via the mucous membranes, by inhalation or ingestion. Hazardous waste must be packaged, transferred and disposed of properly to protect both people and the environment. To this end, the work is aimed at doing microbiological studies of waste dumpsite in Abia State University Teaching Hospital (ABSUTH), Aba.
| Materials and Methods | |  |
Study area
The study area was ABSUTH, Aba. The University Teaching Hospital was chosen because of the large population of people that visit the hospital. This population figure has a direct relationship with the volume of waste generated as stipulated, Nigerian Environmental Study Action Team (1991).
Passive air sampling: Settle plates
Passive air sampling was performed using settle plates. Petri dish More Detailses containing a solid media were left open to air for a given period of time (for 1 h, 1 m from the floor, at least 1 m away from walls or any obstacle). Microbes carried by inert particles fall onto the surface of the nutrient, with an average deposition rate of 0.46 cm/s as reported by. [3] After incubation at 36°C ± 1°C they colonies grow in a number proportional to the level of microbial contamination of the air.
Collection and micro-analysis of hospital soil samples
The samples for soil microbiological analysis were single samples of soil sediment underlying SW were collected from the common dumping site located in the hospitals and soil adjacent to the dumpsites were also collected. All samples were taken from a depth of 0.9 to 30 cm.
The samples were transferred to polythene bags approved by Nigeria's Federal Environmental Protection Agency. These were sealed and transported to the Microbiology Research Laboratory of Abia State University, Uturu for analysis while the remains were stored in a deep freezer (–20°C) for further microbiological analysis.
Collection and micro-analysis of hospital wastewater drippings
The wastewater samples were collected from the drippings of the SW dumpsite of the University Teaching Hospitals in Southeast Nigeria.
Preparation of media for microbiological analysis
The media used for the isolation of microorganisms include nutrient agar, MacConkey agar, sabroud dextrose agar, potato dextrose agar, mannitol salt agar, eosin methylene blue agar, Salmonella More Details-Shigella agar, blood agar and nutrient broth. The different media used in isolation were prepared according to the manufacturer's instructions.
Isolation of microorganism from the collected samples
The microbiological parameters monitored include total viable aerobic counts to isolate heterotrophic bacterial and fungal, total coliform counts (TCCs), according to the methods of. [4]
Bacteria were isolated and characterized using cultural identification, morphological identification using Gram staining reaction and other biochemical tests which include; catalase, methyl red, voges proskauer, nitrate reduction test, starch hydrolysis, gelatin liquefaction test, coagulase, indole, motility, oxidase, urease, triple sugar iron agar and sugar fermentation as described by Bergey's manual of determinative bacteriology. [5],[4]
While fungi was isolated using the growth rate, colonial morphological features and microscopic morphological features. The color of aerial hyphae and substrate hyphae was observed and staining procedure as described by. [5],[6]
Molecular analysis
DNA extraction was performed at the anaerobe laboratory, molecular biology and biotechnology division, Nigerian Institute of Medical Research Yaba Lagos. Methodology was based on polymerase chain reaction (PCR) and metagenomics analysis. While sequencing analysis was done at Inqaba Biotechnology Pty, South Africa by using Sanger (dideoxy) sequencing technique to determine the nucleotide sequence of the specific microorganism isolated using automated PCR cycle-Sanger Sequencer™ 3730/3730XL DNA analyzers from applied biosystems. [7],[8]
Drug sensitivity pattern of bacteria isolates
The antibacterial activity of the different concentrations of various antibiotics; tetracycline (30 μg), ampicillin (10 μg), chloramphenicol (30 μg), ciprofloxacin (30 μg), cotrimaxazole (25 μg), were determined using disc diffusion method.
Statistical analyses
Data generated from this study was analyzed using IBM SPSS statistical software, Chi-square test and analysis of variance. The variables was expressed in mean and standard deviation. A P < 0.05 was considered statistically significant.
| Results | |  |
The result of the total microbial count of air samples within the waste dumpsite and reception unit of ABSUTH, Aba revealed that the total viable count (TVC) was 182 ± 0.5 air within waste dumpsite (AW) and 70 ± 2.0 air within reception unit (AR), the TCC was 17 ± 0.2 (AW) and 5 ± 1.0* (AR), the total staphylococcal count (TSC) was 8 ± 0.1 (AW) and 3 ± 0.5* (AR) while the total fungal count was 60 ± 0.4 (AW) and 42 ± 0.7 (AR), the variations in the results differ significantly at P < 0.05 [Table 1].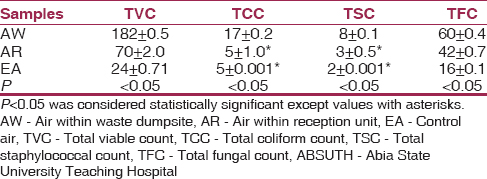 | Table 1: Total microbial count of air samples within the waste dumpsite and reception unit of ABSUTH, Aba
Click here to view |
Total microbial count of soil samples underlying SW and soil adjacent to the dumpsite of ABSUTH, Aba showed that the TVC was 3.0 ± 1.7 × 10 8 cfu/g and 4.1 ± 0.4 × 10 7 cfu/g for soil underlying SW and soil adjacent to dumpsite (SD), which differ significantly at P < 0.05 when compared with the control soil (ES), 3.2 ± 0.8 × 10 5 cfu/g. The TCC was 1.6 ± 0.1 × 10 4 cfu/g SW to 2.3 ± 0.2 × 10 4 cfu/g SD and differs significantly (P > 0.05) with the control (1.0 ± 1.0 × 10 4 cfu/g), the TSC 1.9 ± 0.4 × 10 2 cfu/g (SW) and 2.8 ± 0.1 × 10 2 (SD) and do not differ significantly (P > 0.05) with the control (2.1 ± 0.1 × 10 2 cfu/g) while the total fungal counts showed a significant difference when compared with the control (5.3 ± 0.6 × 10 6 cfu/g) and was 6.8 ± 0.3 × 10 7 cfu/g, 7.8 ± 0.5 × 10 7 for soil underlying SW and SD, respectively [Table 2].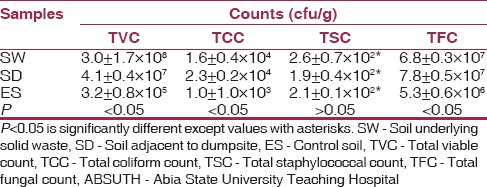 | Table 2: Total microbial count of soil samples underlying solid waste and soil adjacent to the dumpsite of ABSUTH, Aba
Click here to view |
The variations in microbial counts (TVC 6.9 ± 1.7 × 10 7 , TCC 3.5 ± 1.0 × 10 3 , TSC 1.8 ± 0.6 × 10 2 and TFC 7.4 ± 0.1 × 10 5 ) of the dripping samples from the waste dumpsite differed significantly (P < 0.05) when compared with the control which showed no growth [Table 3].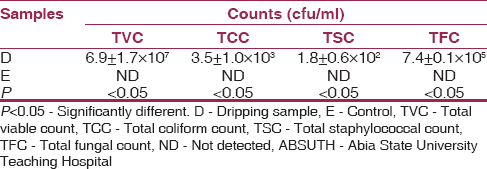 | Table 3: Total microbial count of dripping samples from the waste dumpsite of ABSUTH, Aba
Click here to view |
The microorganisms isolated from the various samples and the controls include, Erwinia herbicola, Serratia species, Staphylococcus saprophyticus, Enterococcus faecalis, Staphylococcus aureus, Streptococcus species, Escherichia More Details coli, Salmonella sp., Pseudomonas aeruginosa, Proteus mirabilis, Staphylococcus epidermidis, Corynebacterium species, Neiseria species, Actinomycetes isreali, Klebsiella aerogenes, Shigella species, Bacillus species, Aspergillus flavus, Candida utilis, Aspergillus fumigatus, Aspergillus niger, Rhodotorula glutinis, Penicillium spp. and Microsporium distortum [Table 4].
[Table 5] shows the susceptibility profile of the bacterial isolates from the hospital wastes, this revealed that ciprofloxacin and augumentin produced the highest percentage inhibition of 33 to 100% against all the bacterial isolates except A. isreali which was resistant to Augumentin. Gentamycin was sensitive against Klebsiella spp., Serratia spp. and S. aureus, marked resistance was recorded against Bacillus spp.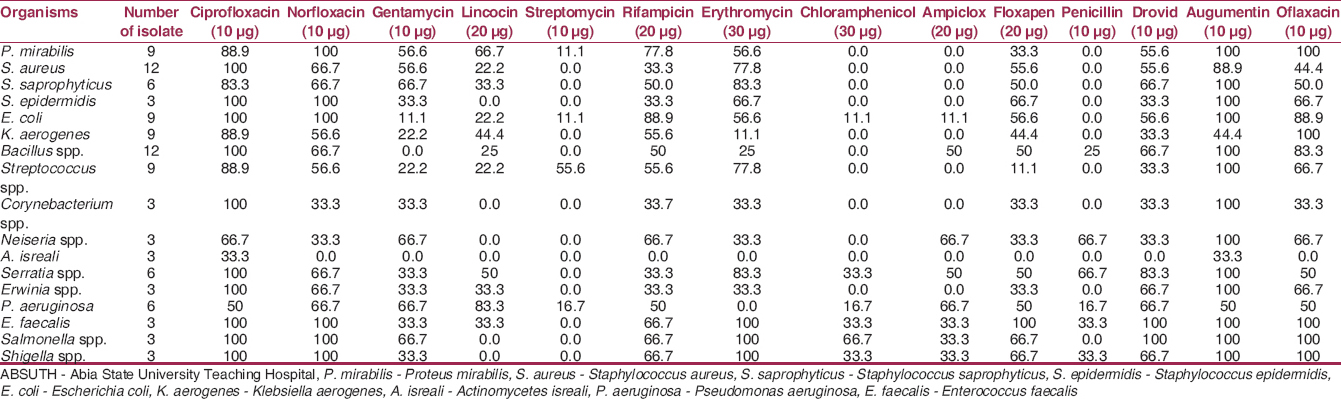 | Table 5: Percentage antibiotic susceptibility pattern of bacterial isolates from ABSUTH
Click here to view |
[Table 6] shows the plasmid profile of bacteria and fungi isolates from waste dumpsite and control where most of the isolates had no plasmid except for E. coli which produced equal size of 23 kb of plasmid.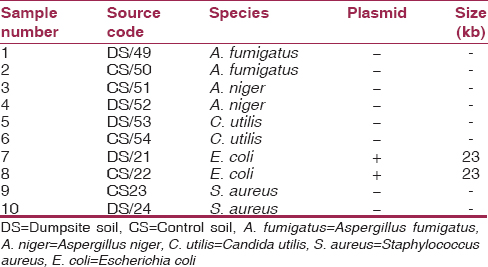 | Table 6: The plasmid profile of most encountered bacteria and fungi isolates from waste dumpsite and control
Click here to view |
[Figure 1] shows the Agarose gel of plasmid DNA showing the presence (lanes 9 and 10) and absence (lanes 1-8) of Plasmid bands Lane M.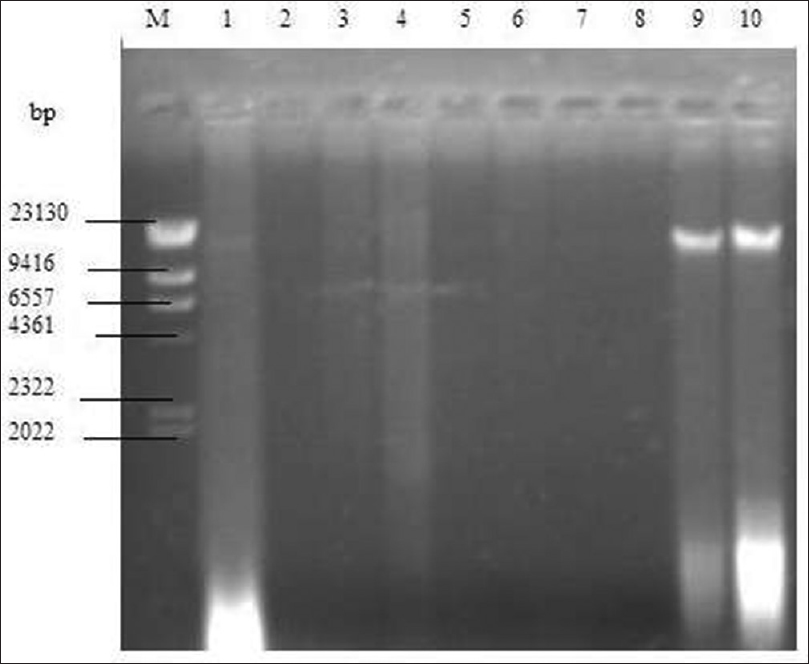 | Figure 1: Agarose gel of plasmid DNA showing the presence (lanes 9 and 10) and absence (lanes 1-8) of plasmid bands lane M: hind III DNA standard marker, lane 1 -Aspergillus fumigatus dumpsite soil (DS)-49, lane 2 -A. fumigatus control soil (CS)-50, lane 3 -Aspergillus niger CS-51, lane 4 -A. niger DS-52, lane 5 -Candida subtilis DS-53, lane 6 -C. subtilis CS-54, lane 7 -Staphylococcus aureus Cb-23, lane 8 -S. aureus DS-24, lane 9 -Escherichia coli DS-21, lane 10 -E. coli CS-22
Click here to view |
| Discussion | |  |
A hospital produces waste by giving their services to the patients. This waste can be produced directly in combination with the service (e.g. injection) or in the upstream (e.g. blood or urine cultures in the laboratory) or downstream (vaccine) process. One kind of typical diseases treated in hospitals is infectious diseases. By the services for known or unknown infectious patients, waste can be the source of an infectious agent
In the present study, microbiological studies of waste dumpsite in ABSUTH, Aba was evaluated. The results revealed that the TVC was 182 ± 0.5 (AW) and 70 ± 2.0 (AR), the TCC was 17 ± 0.2 (AW) and 5 ± 1.0* (AR), the TSC was 8 ± 0.1 (AW) and 3 ± 0.5* (AR) while the total fungal count was 60 ± 0.4 (AW) and 42 ± 0.7 (AR), the variations in the results differ significantly at P < 0.05. However, the microbial load of the air around the hospital waste dumpsite showed high microbial load when compared with the control sample and the air within the teaching hospital reception. This observation disagreed with the results of an earlier study [9] which reported low microbial load in intramural hospital environment and agrees with another study [10] which reported high bacterial load in the extramural environment. This fact could be attributed to the dumpsite area encouraging the growth and proliferation of microorganisms which are easily carried by air. Also, the dumpsite environment could have influenced the count obtained from the reception area directly or indirectly, especially as the dumpsite is close to the teaching hospital and could have a direct effect on those attending or working there. This could lead to increase in disease condition or increase in hospital acquired infections.
Total microbial count of soil samples underlying SW and soil adjacent to the dumpsite of ABSUTH, Aba showed that the TVC was 3.0 ± 1.7 × 10 8 cfu/g and 4.1 ± 0.4 × 10 7 cfu/g for soil underlying SW and SD, which differ significantly at P < 0.05 when compared with the ES, 3.2 ± 0.8 × 10 5 cfu/g. The TCC was 1.6 ± 0.1 × 10 4 cfu/g (SW) to 2.3 ± 0.2 × 10 4 cfu/g (SD) and differs significantly (P > 0.05) with the control (1.0 ± 1.0 × 10 4 cfu/g), the TSC 1.9 ± 0.4 × 10 2 cfu/g (SW) and 2.8 ± 0.1 × 10 2 (SD) and do not differ significantly (P > 0.05) with the control (2.1 ± 0.1 × 10 2 cfu/g) while the total fungal counts showed a significant difference when compared with the control (5.3 ± 0.6 × 10 6 cfu/g) and was 6.8 ± 0.3 × 10 7 cfu/g, 7.8 ± 0.5 × 10 7 for soil underlying SW and SD, respectively. An earlier study [11] noted that high counts of bacterial load reflected the level of pollution in the environment, which is an indication of the amount of organic matter present. However, the high count observed in the study is of public health importance. These organisms can easily be carried into the hospitals environment by foot or other means which could increase the risk of infection. Also, they can be washed down by rain into the underground water of the hospitals leading to water contamination and spread of water borne diseases.
The variations in microbial counts (TVC 6.9 ± 1.7 × 10 7 , TCC 3.5 ± 1.0 × 10 3 , TSC 1.8 ± 0.6 × 10 2 and TFC 7.4 ± 0.1 × 10 5 ) of the dripping samples from the waste dumpsite differed significantly (P < 0.05) when compared with the control which showed no growth. This is an indication that the water that drips out of the hospital waste dumpsite are contaminated with potential pathogenic microorganisms which could pose a potential health risk when such drippings contaminate public water sources. These drippings can also affect the air quality of the hospital environments which is of public health significance.
The microorganisms isolated from the various samples and the controls include, E. herbicola, Serratia species, S. saprophyticus, E. faecalis, S. aureus, Streptococcus species, E. coli, Salmonella sp., P. aeruginosa, P. mirabilis, S. epidermidis, Corynebacterium species, Neiseria species, A. isreali, K. aerogenes, Shigella species, Bacillus species, A. flavus, C. utilis, A. fumigatus, A. niger, R. glutinis, Penicillium spp. and M. distortum. Similar observation has been reported in a study conducted in Benin City, Nigeria. [12] The presence of these organisms in the analyzed samples is of public health significance. This study is in accordance with another study in which 24 hospital waste samples taken from different hospital dump sites were examined. [13] Bacteria isolated at the soil dumpsite and SDs included, respectively, Bacillus sp. (42.86 and 45%), Micrococcus roseus (14.29 and 10%), S. epidermidis (9.52 and 10%), Corynebacterium equi (1.59 and 5%), Bacillus subtilis (4.76 and 5%), Bacillus licheniformis (9.52 and 10%), and Actinomyces istraelii (3.17 and 5%). Fungi isolated included Rhizopus nigricans (27.59 and 18.52%), A. flavus (13.79 and 3.70%), Penicillium rubrum (6.86 and 3.70%), Trichothecium roseum (0 and 3.70%), Penicillium viricadum (6.90 and 0%) A. niger (34.48 and 44.44%), A. nidulans (0 and 11.11%), A. visicolor (3.45 and 3.45%), A. parasiticus (0 and 7.41%), and Microsporum canis (6.9 and 0%). The dumpsites soil recorded higher number of microorganism than the adjacent soil. The pathogens present in the wastes can leak out and contaminate ground water and surface water. Most of these fungi isolated are implicated in various disease conditions and therefore could pose a serious health risk in hospital environments and the public.
The susceptibility profile of bacterial isolates from the samples revealed that ciprofloxacin and augumentin produced the highest percentage inhibition of 33-100% against all the bacterial isolates except A. isreali which was resistant to augumentin.
The presence of plasmid in an organism that is resistant to an antibiotic indicates that that the organism may not be treated by that antibiotic especially if it is the drug of choice for infection. According to the results obtained, it was observed that only the E. coli isolates and its control harbored plasmid with the size of approximately 22,130 kilobase-pairs. The presence of this plasmid can be related with the functions of this organism. The presence of a resistant plasmid in a hospital environment may signify a nosocomial transmission. This is of great concern because management and treatment of infections caused by these resistant plasmid possessing species may lead to therapeutic failure.
| Conclusion | |  |
The high microbial load densities suggests that the hospital wastes in the environment pose a major health and environmental threat, which therefore calls for a proper regulatory system on disposal of hospital waste and there is an urgent need to raise awareness, education and management strategy on medical waste issues to ensure healthy and environmental safety.
| References | |  |
| 1. | Wallace LP, Zaltzman R, Burchinal JC. Where solid waste comes from and where it goes. Mod Hosp 1973;121:91-2.  |
| 2. | De-Roos RL. Environmental concerns in hospital waste disposal. Hospitals 1974;48:120-2.  |
| 3. | Pasquarella C, Pitzurra O, Savino A. The index of microbial air contamination. J Hosp Infect 2000;46:241-56.  |
| 4. | Gerhardt P, Murray RG, Wood WA, Krieg NR. Methods for General and Molecular Bacteriology. Washington DC: ASM Press; 1994. p. 791.  |
| 5. | Sharma S, Chauhan SV. Assessment of bio-medical waste management in three apex government hospitals of Agra. J Environ Biol 2008;29:159-62.  |
| 6. | Cheesbrough M. Water quality analysis. District Laboratory Practice in Tropical Countries. Part 2. United Kingdom: Cambridge University Press; 2003. p. 146-57.  |
| 7. | Russell P. Genetics. San Francisco: Pearson Education Inc.; 2002. p. 187-9.  |
| 8. | |
| 9. | Benediktsdóttir E, Kolstad K. Non-sporeforming anaerobic bacteria in clean surgical wounds - air and skin contamination. J Hosp Infect 1984;5:38-49.  |
| 10. | Apurva K, Pathak W, Karuna SV. Extramural aero-bacteriological quality of hospital environment. Asian J Exp Biol Sci 2010;1:128-35.  |
| 11. | Aluyi AS, Ekhaise OF, Adelusi MD. Effect of human activities and oil pollution on the microbiological and physiological quality of Udu River, Warri, Nigeria. J Appl Sci 2006;6:1214-9.  |
| 12. | Ekhaise FO, Omavwoya BP. Influence of hospital wastewater discharged from University of Benin Teaching Hospital (UBTH), Benin City on its receiving environment. Am Eurasian J Agric Environ Sci 2008;4:484-8.  |
| 13. | Oyeleke SB, Istifanus N. The microbiological effects of hospital wastes on the environment. Afr J Biotechnol 2009;8:6253-7.  |
[Figure 1]
[Table 1], [Table 2], [Table 3], [Table 4], [Table 5], [Table 6]
|