Mind the dbGAP: The Application of Data Mining to Identify Biological Mechanisms
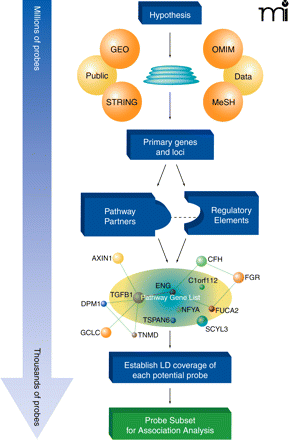
Pathway analysis tools can use biological knowledge to focus the number of genetic markers for association analysis from millions of potential variants to a few thousand tightly focused on the trait under study. The initial hypothesis, based upon a known gene and/or a biological process, forms the basis for selecting Medical Subject Headings (MeSH), which are used to mine both the published literature and the Online Mendelian Inheritance in Man (OMIM) databases. This list is supplemented with genes that are differentially expressed in the setting of the tissue or trait of interest identified from analysis of Gene Expression Omnibus (GEO) microarray datasets. The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) facilitates this process by identifying likely pathway-related genes based on a wide array of knowledge-based interrelationships, including protein interaction, known co-regulation, and comparative genomics. These data-mining steps serve to generate a primary list of genes deemed biologically relevant to the trait under study. Analysis of pathway relationships further builds out the gene list as well as stratifies the gene list by identifying molecular relationships and key partners. All genetic markers present on fixed genotype platforms that are also contained within or nearby pathway genes (e.g., in upstream or downstream regulatory sequences) are then identified. Pathway SNPs that provide similar overall information content by virtue of being in high LD are removed to focus the list of markers and reduce the overall testing penalty. Finally, the refined list of Pathway SNPs is used to test genotype-phenotype associations in novel or dbGAP-derived datasets. ENG, endoglin; TGFB1, transforming growth factor–beta 1; DPM1, dolichyl-phosphate mannosyltransferase polypeptide 1; GCLC, glutamate-cysteine ligase; TNMD, tenomodulin; TSPAN6, tetraspanin 6; SCYL3, SCY1-like 3; FUCA2, fucosidase, alpha-L- 2; FGR, Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog; CFH, complement factor H; AXIN1, axin 1; C1orf112, chromosome 1 open reading frame 112; NFYA, nuclear transcription factor Y, alpha.



