Na+-K+–ATPase-Mediated Signal Transduction: From Protein Interaction to Cellular Function
- Zijian Xie1,2 and
- Ting Cai1
Abstract
The Na+-K+-ATPase, or Na+ pump, is a member of the P-type ATPase superfamily. In addition to pumping ions, Na+-K+-ATPase is engaged in assembly of multiple protein complexes that transmit signals to different intracellular compartments. The signaling function of the enzyme appears to have been acquired through the evolutionary incorporation of many specific binding motifs that interact with proteins and ligands. In some cell types the signaling Na+--ATPase and its protein partners are compartmentalized in coated pits (i.e., caveolae) the plasma membrane. Binding of ouabain to the signaling Na+-K+-ATPase activates the cytoplasmic tyrosine kinase Src, resulting in the formation of an active “binary receptor” that phosphorylates and assembles other proteins into different signaling modules. This in turn activates multiple protein kinase cascades including mitogen-activated protein kinases and protein kinase C isozymes in a cell-specific manner. It also increases mitochondrial production of reactive oxygen species (ROS)and regulates intracellular calcium concentration. Crosstalk among the activated pathways eventually results in changes in the expression of a number of genes. Although ouabain stimulates hypertrophic growth in cardiac myocytes and proliferation in smooth muscle cells, it also induces apoptosis in many malignant cells. Finally, the signaling function of the enzyme is also pivotal to ouabain-induced nongenomic effects on cardiac myocytes.
Foxglove (Digitalis purpurea), from which the cardiac glycoside compounds digitalis and digitonin are derived.
Introduction
The Na+ -K+ –ATPase (EC 3.6.3.9), or Na+ pump, is an energy-transducing ion pump first described by Skou in 1957 (1) . Since then, the structure, the pumping function, and the regulation of the enzyme have been well characterized (2 –4) . In addition to pumping ions across the membrane, the enzyme, composed of both catalytic α and regulatory β subunits, functions as a receptor for cardiotonic steroids (CTS) (5 –8) . Because CTS inhibit Na+ -K+ –ATPase, the physiological and pharmacological functions of CTS were thought secondary to their effects on intracellular ion concentrations. However, the concentration of endogenous ouabain, even under pathological conditions, is not high enough to cause changes in global cytosolic ion concentrations. This fact has prompted many laboratories to test whether ouabain-liganded Na+ -K+ –ATPase can generate secondary messages independent of its pumping function (9 –15) . Subsequently, several mechanisms and pathways by which Na+ -K+ –ATPase transmits the ouabain signal to different intracellular compartments have been discovered (16 –22) . It is the purpose of this review to provide a broad overview of Na+ -K+ –ATPase research with detailed discussions on the mechanisms and biological significance of Na+ -K+ –ATPase-mediated signal transduction.
Protein–Protein Interactions and Na+-K+-ATPase-Mediated Signal Transduction
Binding of CTS to the Na+ -K+ –ATPase affects gene expression and cell growth (23 , 24) . Early studies indicated that the enzyme interacts with many soluble and membrane proteins (25) ; however, only in recent years has research on Na+ -K+ –ATPase revealed that interactions of Na+ -K+ –ATPase with other proteins not only are important for regulation of pumping function, but also make it possible for the enzyme to function as a signal transducer (25) .
Evolving of Protein-Binding Motifs and Signal Transduction
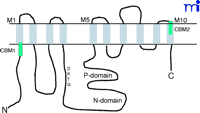
P-type ATPases are expressed in all living organisms (26) . The primary structures of various P-type ATPase catalytic subunits contain eight conserved motifs solvated in the cytoplasm, where ATP binding and hydrolysis occur (26 , 27) . The primary function of Na+ -K+ –ATPase is to transport ions across the cell membrane; the signaling function of Na+ -K+ –ATPase most likely arose from the separate evolution of specific binding motifs. Indeed, computational analyses have revealed that the human Na+ -K+ –ATPase α subunit contains multiple well-characterized protein-binding motifs (Table 1⇓), all of which are located outside of the eight conserved core regions of P-type ATPases (26) . Because the interaction between Na+ -K+ –ATPase and caveolins—the protein constituents of caveolae located on the plasma membrane—plays a role in Na+ -K+ –ATPase-mediated signal transduction (28 , 29) , the caveolin-binding motif (CBM), found on the Na+ pump, has been used as a model for further analysis, leading to the following observations. First, human Na+ -K+ –ATPase α1 subunit contains two potential CBMs (Table 1⇓), one of which is in the cytosolic N-terminal domain near the first transmembrane region (M1), whereas the other CBM is located at the extracellular side of M10 (Figure 1⇓). According to the phylogenetic analyses of the Na+ pump (26, 27) , the N-terminal CBM of the Na+ pump’s α subunits was acquired quite early during evolution, as were those found in the epithelial growth factor receptor (EGFR) (Table 2⇓). It first appeared in one of the mutated Na+ -K+ –ATPases in C. elegans (i.e., Eat-6) and is conserved in Drosophila (26) (Table 1⇓). Second, the appearance of the N-terminal CBM correlates well with the occurrence of the domain for ouabain binding (27) , suggesting that ouabain regulates not only the ATPase activity but also the interactions of the enzyme with other proteins. In other words, evolutionary acquisition of this domain added a new function to Na+ -K+ –ATPase so that the enzyme can transduce the extracellular ouabain signal. Third, unlike CMBs in other proteins, the CBMs of the Na+ pump’s α subunits are located at sites far away from the catalytic P and N domains (Figure 1⇓). Thus, it is conceivable that binding to caveolin will not affect the ion-pumping function of the enzyme. On the other hand, this interaction could serve to concentrate the enzyme and other signaling proteins into caveolae, resulting in the formation of a large signaling complex or signalosome (Figure 2⇓).
Schematic presentation of the α1subunit of Na+-K+-ATPase. The ten transmembrane domains are marked as M1 to M10. The third intracellular loop, connecting the M4 and M5, contains both P (phosphorylation) domain and N (nucleotidebinding) domain. The green bars indicate caveolin-binding motifs (CBMs).
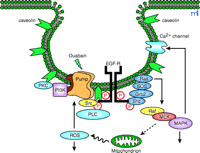
Schematic presentation of Na+-K+-ATPase signalosome. The Na+-K+-ATPase (pump) is preassembled with its partners in caveolae; ouabain binding to the pump activates the signalosome, and transduces signals via multiple pathways (see text for details). EGFR, epithelial growth factor receptor; PKC, protein kinase C; PI3K, phosphoinositide 3’ kinase; Grb2, growth factor receptor-bound protein 2; Sos, son of sevenless; Shc, src homology collagenlike protein; PLC, phospholipase C; MAPK, mitogen-activated protein kinase; MEK, MAPK-ERK activating kinase; ROS, reactive oxygen species.
Na+-K+–ATPase Signalosome
As illustrated in Figure 2⇑, the Na+ -K+ –ATPase interacts with multiple signaling proteins to transmit ouabain signals (14 –22 , 30 –33) . Prior studies also demonstrate the interactions of the Na+ -K+ –ATPase with the Na+ pump’s γ subunit (FXYD2), phospholemman (FXYD1), and other FXYD proteins in a cell-specific manner (34 , 35) . The FXYD proteins share a signature sequence of six conserved amino acids that include the FXYD motif, where F, Y, and D represent amino acid residues of phenylalanine, tyrosine, and aspartic acid, respectively (34) . The Na+ pump also binds to ankyrin and adducin (36 –38) . Interestingly, polymorphisms in the sequence of adducin correlate with hypertension in both humans and Milan rats (37) . These observations demonstrate the importance of protein–protein interactions in the regulation of both the ion-pumping and signal-transducing functions of the Na+ -K+ -ATPase, and have prompted several laboratories to isolate the intact Na+ -K+ –ATPase signalosome so that the composition of the Na+ pump’s interacting proteins can be identified using newly developed proteomic approaches (14 , 22 , 39 –43 ). For example, using ouabain-affinity column, Yingst et al. demonstrated the feasibility of purifying Na+ -K+ -ATPase from various types of cells and tissue samples (41) . In principle, when experimental conditions are controlled properly, this method could be used to isolate the intact signalosome of the enzyme. In addition, Caplan’s laboratory proved GST (glutathione S -transferase) pull-down assays to be a useful tool for purification of the intact Na+ -K+ -ATPase signalosome from cultured cells (42) . We also have performed experiments to test the feasibility of using a combination of protein purification, GST pull-down, immunoprecipitation, and matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) analysis to define the composition of Na+ -K+ -ATPase signalosome (43) . From the identified proteins a picture emerges of a signaling module responsible for the ouabain-mediated regulation of intracellular calcium ([Ca2+ ]i ). The transducer of this module is the Na+ -K+ –ATPase. The messengers are generated by the interaction of the enzyme with the tyrosine kinase Src and phospholipase C–γ (PLC-γ ), and transmitted to the effector IP3 receptor (IP3R) (43) . The involvement of IP3R in ouabain-induced calcium oscillation has been reported previously (15) . We believe that the collective efforts of many laboratories in the field will eventually map the composition of Na+ -K+ –ATPase signalosome, which may reveal how Na+ -K+ –ATPase is organized spatially and temporally with its partners to transmit signals. This information will also allow us to test whether Na+ -K+ –ATPase participates in the events activated by growth factors and other hormones; there is evidence that the enzyme plays a scaffolding role in other stimuli-induced signal transduction (e.g., dopamine-induced activation of PI3K) (39) . Because protein–protein interactions are important for regulating the pumping function of the enzyme (35 , 39) , the defined composition of the Na+ -K+ –ATPase signalosome under specific conditions may yield new insights into the mechanism by which Na+ -K+ –ATPase is regulated by various stimuli in a cell-specific manner.
Interaction of Na+-K+–ATPase with Src
The binding of ouabain to the Na+ -K+ –ATPase causes rapid activation of Src family kinases in many different types of cells, including cardiac myocytes, smooth muscle, and kidney epithelial cells (14 , 21 , 22) . The kinases are 52–62-kDa membrane-associated non-receptor tyrosine kinases that regulate various signal transduction pathways (44) , and their catalytic activity is controlled by tyrosine phosphorylation and protein–protein interactions (44 –46) . Phosphorylation of Tyr529 on Src holds the kinase in an inactive conformation through an intramolecular interaction with its Src homology (SH) 2 (SH2) domain, whereas phosphorylation of Tyr418 activates Src by disrupting the intramolecular interaction and creating the substrate-binding site. The complexities of the interactions that hold Src in an inactive conformation suggest that there are many mechanisms for activating the kinase (45 , 46) .
Because the effects of ouabain on Src are independent of changes in intracellular ion concentrations (14 , 21 , 22) , we proposed that Src might be activated by ouabain-induced changes in the interactions between Na+ -K+ –ATPase and Src. This hypothesis is supported by the following observations. First, ouabain stimulates Tyr418 phosphorylation but has no effect on Tyr529 phosphorylation of Src in different types of cells (22) . In addition, when the α1 subunit of the Na+ pump is immunoprecipitated from both ouabain-treated and control LLC-PK1 cells, ouabain increases the binding of Src to the Na+ -K+ –ATPase signaling complex in a dose- and time-dependent manner (22) . Third, GST pull-down assays show that the solubilized Na+ -K+ –ATPase binds to GST-Src in a dose-dependent manner (47) , and that interaction involves both kinase domain and the SH3 and SH2 domains of Src. Fourth, purified Src interacts with a GST fusion protein encompassing the large central loop between transmembrane helices M4 and M5 of the α1 subunit of the Na+ pump. Several recent studies indicate that this domain may be involved in stabilizing the α –α subunit interactions of the Na+ pump as well as the interaction of the α subunit with other proteins (48 –50) . In short, the above observations clearly demonstrate that Src interacts directly with Na+ -K+ –ATPase via multiple domains. However, it remains unclear how ouabain regulates these interactions and activates Src.
The “Binary Receptor” Relays Ouabain Signals from Na+-K+–ATPase to Other Signaling Cascades
Because Src interacts with the Na+ pump directly, we suggest that the Na+ -K+ –ATPase–Src complex behaves as a “binary” receptor (Figure 2⇑). Once the receptor is activated by ouabain, it indirectly affects the phosphorylation of downstream proteins that are associated with or are proximal to the receptor. Indeed, when ouabain-treated LLC-PK1 cell lysates are immunoprecipitated with α1 -specific antibody, Src-dependent tyrosine phosphorylation of multiple Na+ -K+ –ATPase-associated proteins is observed (47) .
The activated EGFR is a critical element in the signal transduction networks of cytokines, H2 O2 , and those pathways utilizing G protein–coupled receptors (51) . Src family kinases can couple receptors that lack intrinsic kinase activity to receptor tyrosine kinases (RTKs), such as EGFR. Indeed, as depicted in Figure 2⇑, activated Src transactivates EGFR, which in turn recruits adapter protein Shc to relay the initial ouabain signal to the Ras–Raf–MAPKs cascade (14 , 22) . Furthermore, Src activation is also required for ouabain-induced stimulation of several other pathways including increased production of reactive oxygen species (ROS) and activation of PKC isozymes. Taken together, these findings support the proposal that the activation of the Na+ -K+ –ATPase–Src complex is the initial critical step that relays the signal emanating from the interaction of ouabain with the enzyme to the EGFR and other downstream pathways.
Although the above findings reaffirm the importance of protein–protein interaction in Na+ -K+ –ATPase-mediated signal transduction, it is important to note that large changes in intracellular ion concentrations can alter different ion concentration–sensitive signaling events (23 , 52 , 53) . Furthermore, because local cytosolic Na+ concentrations could be affected when a small percentage of Na+ pumps is inhibited by low doses of ouabain (54) , the impact of changes in local ion concentration on ouabain-activated signaling events should also be considered.
Are Na+-K+–ATPase Signalosomes Compartmentalized?
Because the Na+ -K+ –ATPase has to interact with Src, EGFR, and other proteins to transmit the ouabain signal, it is possible that the signaling enzyme may have to be preassembled with its signaling partners in caveolae. Caveolae are membrane microdomains that were first identified as flask-shaped vesicular invaginations of plasma membrane enriched in cholesterol, glycosphingolipids, and sphingomyelin (55 , 56) . Caveolins are 21–24-kDa membrane-associated scaffolding proteins, with multiple cellular functions, that typify caveolae (55) . Mammals express three different caveolin genes that encode five different protein isoforms, the expression and distribution of which are tissue-specific (56) . Caveolins directly interact with cholesterol and many signaling proteins including receptors, Src family kinases, and adapter proteins. Recent studies have indicated that many of these interactions are mediated through the binding of caveolin scaffolding domains to caveolin-binding motifs of the target proteins. Two potential caveolin-binding motifs exist in the α1 subunit of Na+ -K+ –ATPase (Table 1⇑). Confocal imaging shows that Na+ -K+ –ATPase colocalizes with caveolins on plasma membrane in cardiac myocytes and other types of cells (28 , 29) . Immunoprecipitation using caveolin-1–specific antibody-coated magnetic beads confirmes that the Na+ pump is associated with caveolae, Src, and other signaling proteins. GST pull-down assays reveals that Na+ -K+ –ATPase binds to the N terminus of caveolin-1 (29) . Significantly, ouabain appears to regulate the interaction of caveolin-1 with Na+ -K+ –ATPase and induces the formation of the Na+ -K+ –ATPase–Src–caveolin-1 complex. Ouabain also stimulates tyrosine phosphorylation of caveolin-1 in LLC-PK1 cells, and caveolar p42/44 MAPKs in cardiac myocytes (28 , 29) . Furthermore, removal of cholesterol significantly reduces ouabain-induced activation of extracellular-regulated protein kinases (ERKs) in LLC-PK1 cells which is reversible after cholesterol repletion in these cells. These findings support the notion that ouabain activates the caveolar Na+ -K+ –ATPase to initiate different signal transduction pathways (29) .
Endogenous Ouabain and Other Na+-K+–ATPase Ligands
CTS consist of a group of chemicals that specifically bind to Na+ -K+ –ATPase. They include plant-derived digitalis drugs such as digoxin and ouabain, and vertebrate-derived aglycones such as bufalin and marinobufagenin. Although they have been considered only as drugs for a long time, there is now ample evidence to indicate that some of these compounds can function as steroid hormones (57 –62) . Recent studies have identified both ouabain and marinobufagenin as endogenous steroids whose production and secretion are regulated by multiple pathological or physiological stimuli including angiotensin II and epinephrine (55 , 58, 61 , 62) . In addition to CTS, changes in intracellular Na+ or extracellular K+ can cause conformational changes in the enzyme. These changes affect not only the enzyme conformation, but also membrane potential and other ion transporter–related activities; therefore, they could trigger other indirect signaling events mediated by ouabain. Indeed, although lowering of extracellular K+ does activate protein kinases and raise intracellular calcium in cardiac myocytes (21) , it does not mimic the effects of ouabain on smooth muscle and kidney epithelial cells (14 , 15) . ROS are another group of molecules whose interaction with the Na+ pump can cause a conformational change in Na+ -K+ –ATPase (63 , 64) . This is of particular interest because both endogenous ouabain and ROS concentrations are elevated in several diseases. If ROS stimulates the signaling function of Na+ -K+ –ATPase, then a positive amplification loop would be established because activation of the signaling function of Na+ -K+ –ATPase stimulates the mitochondrial production of ROS (12 , 19 , 65) (Figure 2⇑).
Regulation of Cellular Function by Na+ Pump Signals
Binding of ouabain to Na+ -K+ –ATPase affects multiple cellular functions. The effects of ouabain on gene expression, cell attachment, formation of tight junctions and induction of polarity, modification of immune response, protein trafficking, and ion fluxes have been well documented (20 , 66 –71) . Although some of ouabain’s effects are clearly mediated by Na+ -K+ –ATPase signaling only (67 , 69 –71) , other effects involve both the pumping and signaling functions of the enzyme (68) . Because of space limitations, we will only discuss the relationship between the signaling function of Na+ -K+ –ATPase and the effects of ouabain on cell growth and cardiac function.
Regulation of Cell Growth
As early as the 1960s, ouabain was found to inhibit the mitogen-induced differentiation and proliferation of lymphocytes as well as the growth of malignant cells (24 , 72) . Significantly, there is evidence that administration of digoxin appears to reduce the mortality rate in breast cancer patients (73) . In the laboratory, ouabain has been used in assays to identify mutagenized cell clones that express ouabain-resistant Na+ -K+ –ATPase mutants (74 , 75) . More recently, ouabain and related CTS were found to cause either apoptosis or hypertrophic/proliferative growth in a cell type–specific manner (76 –87) .
Hypertrophic or Proliferative Growth
Cardiac tissue
In addition to their effects on cardiac contractility, digitalis drugs appear to promote cardiac hypertrophy. Based on clinical observations, Christian has argued that in patients with heart disease but not heart failure, digitalis retards cardiac enlargement and delays the appearance of symptoms of cardiac insufficiency (88) . This observation prompted investigation into how digitalis affects growth and hypertrophy in the heart. Rather confusingly, early studies either confirmed that digitalis prevented overload-induced cardiac hypertrophy, showed that digitalis induced or amplified cardiac hypertrophy, or suggested that digitalis had no effect on cardiac growth (89 –92) . In retrospect, these apparent inconsistencies are not surprising, because overload-induced hypertrophy of the heart in an intact animal arises from a complex interplay of changes in a multitude of mechanic and neurohumoral stimuli. Treatment of intact animals with digitalis not only adds another growth stimulus that acts on myocardium directly, but through its well-established neural and vascular effects, can also exacerbate existing pathophysiological stimuli. Furthermore, the different ouabain sensitivities of the experimental animals add another layer of complexity to these studies. Consequently, we have used cultured cardiac myocytes as a model to demonstrate that ouabain stimulates hypertrophic growth in cardiac myocytes (85 –87) . Like other hypertrophic stimuli, ouabain regulates transcription of several hypertrophic marker genes in cardiac myocytes (17 , 85) . Furthermore, the effects of ouabain on cardiac gene expression and growth depend on the signaling function of Na+ -K+ –ATPase and involve crosstalk among different signaling pathways (18 , 19) . A recent study (93) supports our conclusion and demonstrates a correlation between plasma ouabain concentration and increases in left-ventricular mass in human subjects; this correlation exists even when the blood pressure is accounted for.
Cell lines
In addition to stimulating the hypertrophic growth of cardiac myocytes, ouabain also increases the proliferation of several different cell lines. Exposure of canine vascular smooth muscle cells to ouabain concentrations that cause no detectable inhibition (< 5%) of pump activity activates the signaling function of Na+ -K+ –ATPase (14 , 84) . Concomitantly, ouabain also stimulates proliferation in these cells. These findings are significant in view of recent evidence whereby infused ouabain caused significant increases in blood pressure (60) . Because these ouabain concentrations (nM) can affect the signaling (14 , 15) but not bulk cytosolic ion concentrations, the growth effects may explain some of the actions of ouabain on blood pressure.
Apoptosis
Several CTS in nontoxic concentrations can induce apoptosis in different malignant cell lines (76 –82) . Bufalin, for example, induces apoptosis in leukemia cells and human skin squamous carcinoma cells (76 , 81) . Furthermore, other digitalis-like drugs induce apoptosis or enhance the apoptotic effects of other drugs in human prostate cancer cells (19) . Mechanistically, the apoptotic effects of CTS are related to the downstream effects of activated MAPKs: increased [Ca2+ ]i and ROS, and activation of transcription factors such as activator protein-1 (AP-1), and nuclear factor–κ B (NF-κ B). In view of the complexity of ouabain-activated signal transduction pathways, it is not surprising that so many factors could contribute to ouabain-induced apoptosis.
Regulation of [Ca2+]iand Myocyte Contractility by Ouabain Involves p42/44 MAPKs and ROS
Although it is well established that the effects of digitalis-like drugs on cardiac contractility involve the pumping function of the enzyme and Na+ -Ca2+ -exchanger (NCX) (5 –8) , early work did suggest that the activation of protein kinases and Ca2+ channels also participated (93 , 94) . Our studies indicated that activation of protein tyrosine kinases precedes increases in [Ca2+ ]i in response to ouabain (21) . We have tested whether the signal transducing function of Na+ -K+ –ATPase contributes to nongenomic effects of ouabain on myocytes (13 , 95) and have found the following: First, inhibition of either Src or the small GTPase Ras abolishes ouabain-induced increases in both [Ca2+ ]i and contractility. Second, although activation of p42/44 MAPKs is required for ouabain-induced increases in [Ca2+ ]i , both MAPKs and ROS contribute to ouabain regulation of cardiac contraction (95) . The effects of ouabain on [Ca2+ ]i , at least in part, are due to the activation of L-type Ca2+ channels (93) . This activation is most likely evoked by p42/44 MAPK–mediated phosphorylation of the channels, as demonstrated in neuronal cells (96) . Finally, ouabain regulation of cardiac contractility also involves opening of mitochondrial ATP-sensitive K channels (mitoKATP ) and increases ROS production (95) . Inhibition of mitoKATP by treating cells with 5-hydroxydecanoate significantly reduces ouabain-induced increases in ROS production and contractility. Thus, ouabain regulates cardiac contractility through the activation of at least two major pathways. Activation of p42/44 MAPKs and inhibition of the ion-pumping function of the Na+ -K+ –ATPase by ouabain increase [Ca2+ ]i , whereas opening of mitoKATP stimulates, the production of ROS. Both [Ca2+ ]i and ROS, in turn, work in concert and raise contractility in cardiac myocytes. These findings are supported by a recent report showing that ouabain stimulates contraction in myocytes incubated in Na+ -free medium (97) . These observations need to be replicated in the isolated heart and whole animals; nevertheless, it is important to note the significance of these studies. First, these findings suggest that stimulation of the signaling function of Na+ -K+ –ATPase alone may be sufficient to increase cardiac contractility. Second, they point to the possibility of developing agonists that can affect the signaling, but not pumping, function of the enzyme. Finally, because the opening of the mitoKATP channels participates in ischemic preconditioning (98) , it will be of great interest to test whether digitalis drugs can protect the heart from ischemic injury.
Conclusion
The work of the past few years has demonstrated that Na+ -K+ –ATPase is an important signal transducer. Recent work has begun to define the Na+ -K+ –ATPase signalosome, to identify the signaling modules, and to reveal structural domains that are involved in organization of each module. It is important to emphasize that the above studies are critical for our understanding not only of the events regulated by the signaling Na+ -K+ –ATPase, but also of the enzyme ion-pumping activity in different signaling pathways. Identification of intact in vivo Na+ pump signalosomes will ultimately allow us to test their role in ouabain-induced physiological and pharmacological changes in vitro as well as in transgenic or knock-out animal models. Continued study will also aid us in establishing new molecular targets for the development of therapeutics for cardiovascular diseases as well as cancer.
Acknowledgments
We would like to thank Joe Xie for his assistance in computational analysis of potential binding motifs. This work was supported by NIH grants HL-36573, HL-63238, and HL-67963 awarded by NHLBI.
- © American Society for Pharmacology and Experimental Theraputics 2003
References
- 1.↵
- 2.↵
- 3.
- 4.↵
- 5.↵
- 6.
- 7.
- 8.↵
- 9.↵
- 10.
- 11.
- 12.↵
- 13.↵
- 14.↵
- 15.↵
- 16.↵
- 17.↵
- 18.↵
- 19.↵
- 20.↵
- ↵
- 22.↵
- 23.↵
- 24.↵
- 25.↵
- 26.↵
- 27.↵
- 28.↵
- 29.↵
- 30.↵
- 31.
- 32.
- 33.↵
- 34.↵
- 35.↵
- 36.↵
- 37.↵
- 38.↵
- 39.↵
- 40.
- 41.↵
- 42.↵
- 43.↵
- 44.↵
- 45.↵
- 46.↵
- 47.↵
- 48.↵
- 49.
- 50.↵
- 51.↵
- 52.↵
- 53.↵
- 54.↵
- 55.↵
- 56.↵
- 57.↵
- 58.↵
- 59.
- 60.↵
- 61.↵
- 62.↵
- 63.↵
- 64.↵
- 65.↵
- 66.↵
- 67.↵
- 68.↵
- 69.↵
- 70.
- 71.↵
- 72.↵
- 73.↵
- 74.↵
- 75.↵
- 76.↵
- 77.
- 78.
- 79.
- 80.
- 81.↵
- 82.↵
- 83.
- 84.↵
- 85.↵
- 86.
- 87.↵
- 88.↵
- 89.↵
- 90.
- 91.
- 92.↵
- 93.↵
- 94.↵
- 95.↵
- 96.↵
- 97.↵
- 98.↵
Zijian Xie, PhD , (right), is an Associate Professor of the Departments of Pharmacology and Medicine. Ting Cai, MD, MS , (left), is a Research Associate of the Department of Pharmacology. Both are at the Medical College of Ohio. Address correspondence to ZX. Email: zxie{at}mco.edu FAX: 419-383-2871