
- Institution: Stanford Univ Med Ctr Lane Med Lib/Periodical Dept/Rm L109
- Sign In as Member / Individual
The TASK Family: Two-Pore Domain Background K+ Channels
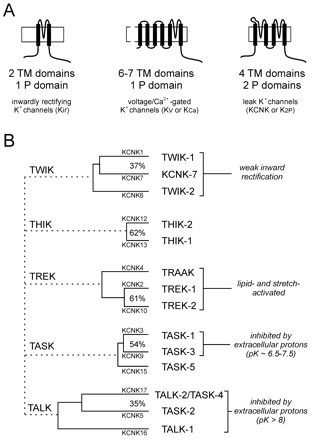
Potassium ion channel gene families. A. Three major K+ channel gene families have been identified: Voltage-gated (Kv) channels have six (or seven) transmembrane segments and one pore domain; inwardly rectifying (Kir) channels present two membrane spanning regions and one pore domain. These channels form as tetramers. The newly identified, weakly rectifying (KCNK) channels are predicted to have four transmembrane regions and two pore domains, each one resembling two Kir channels arranged in tandem. KCNK channels are believed to form as dimers. B. The fourteen known human two-pore-domain K+ channel genes are presented in a phylogenetic tree, and placed into five sub-groups as described (11, 12). Multiple naming schemes for these channels are in current use; however, only two are shown. The Human Genome Organization uses the KCNK designation preceding a number that reflects the order of discovery of each of the genes (as shown here). The International Union of Pharmacology (IUP) adopted a very similar scheme, but replaced the KCNK with a K2P prefix (not shown); the IUP group suggests that a different nomenclature linking clearly related channels will likely be forthcoming (97). One such alternative naming scheme that is in popular use employs a set of acronyms based on salient physiological or pharmacological properties (provided here). TWIK, tandem of P domains in a weak inwardly rectifying K+ channel; THIK, Tandem pore domain halothane-inhibited K+ channel; TREK, TWIK-related K+ channel gene; TRAAK, TWIK-related arachidonic acid-stimulated K+ channel; TASK; TWIK-related acid-sensitive K+ channel; TALK, TWIK-related alkaline pH activated K+ channel.


