
- Institution: Stanford Univ Med Ctr Lane Med Lib/Periodical Dept/Rm L109
- Sign In as Member / Individual
SAR by NMR: Putting the Pieces Together
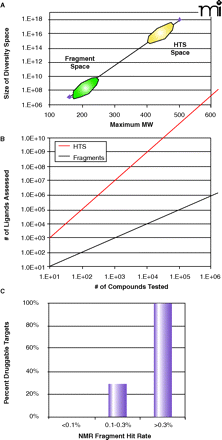
Fragment diversity space. A. A plot of the size of the potential chemical diversity space as a function of the maximum molecular weight for the compound collection (extrapolated from 21 and 22). Note that the y-axis is in log-scale and ranges from 106 to 1018. The approximate regions of diversity space for fragment libraries and conventional HTS collections are denoted by green and yellow ovals, respectively. B. A plot of the virtual number of ligands assessed as a function of the actual number of compounds tested for fragment screening (red line) and conventional high-throughput screening (black line). For HTS, as no linking of the individual hits is proposed, the number of ligands assessed is exactly equal to the number tested. However, in a linked fragment approach, the number of virtual ligands is M*NP, where N is the number of compounds tested, P is the number of independent but proximal subsites on the protein, and M is the number of linking possibilities (23). The example shown here is for (P=2, M=10) as a function of N. C. The observed frequency of being able to inhibit protein targets with potent, non-peptde, non-covalent small molecules as a function of the experimental hit rate from an NMR fragment screen (24).


