|
 
 |
|
ORIGINAL ARTICLE |
|
|
|
| Year : 2013 | Volume
: 19
| Issue : 1 | Page : 14-17 |
| |
Rapid detection of chromosome X, Y, 13, 18, and 21 aneuploidies by primed in situ labeling/synthesis technique
Ashutosh Halder, Manish Jain, Isha Chaudhary
Department of Reproductive Biology, AIIMS, New Delhi, India
| Date of Web Publication | 4-Jun-2013 |
Correspondence Address:
Ashutosh Halder
Department of Reproductive Biology, AIIMS, New Delhi - 110 029
India
 Source of Support: Indian Council of Medical Research and AIIMS, New Delhi, India,, Conflict of Interest: None
DOI: 10.4103/0971-6866.112877

 Abstract Abstract | | |
Aims and Objective: Primed in situ labeling/synthesis (PRINS) technique is an alternative to fluorescent in situ hybridization for chromosome analysis. This study was designed to evaluate the application of PRINS for rapid diagnosis of common chromosomal aneuploidy.
Materials and Methods : We have carried out PRINS using centromere specific oligonucleotide primers for chromosome X, Y, 13, 18 and 21 on lymphocyte metaphase and interphase cells spread. Specific primer was annealed in situ, followed by elongation of primer by Taq DNA polymerase in presence of labeled nucleotides. Finally, reaction was stopped and visualized directly under fluorescent microscope.
Results: Discrete centromere specific signals were observed with each primer.
Conclusion: PRINS seems to be a rapid and reliable method to detect common chromosome aneuploidy in peripheral blood lymphocyte metaphase and interphase cells.
Keywords: Aneuploidies, Chromosome X, Y, 13, 18, and 21, PRINS
How to cite this article:
Halder A, Jain M, Chaudhary I. Rapid detection of chromosome X, Y, 13, 18, and 21 aneuploidies by primed in situ labeling/synthesis technique. Indian J Hum Genet 2013;19:14-7 |
How to cite this URL:
Halder A, Jain M, Chaudhary I. Rapid detection of chromosome X, Y, 13, 18, and 21 aneuploidies by primed in situ labeling/synthesis technique. Indian J Hum Genet [serial online] 2013 [cited 2016 May 24];19:14-7. Available from: http://www.ijhg.com/text.asp?2013/19/1/14/112877 |
 Introduction Introduction | |  |
Molecular cytogenetics is an emerging branch with potentials in clinical practice in particular prenatal-postnatal cytogenetics. Researchers working in molecular cytogenetics field are facing difficulties with the cost and availability of fluorescent in situ hybridization (FISH) probes in particular, centromere specific probes for chromosome 21, chromosome 13, etc., leading to limited applications and reach. Primed in situ labeling/synthesis (PRINS) technology can overcome some of the problems of FISH through alternative rapid and cost-effective procedure. PRINS is a method of target DNA sequence detection and localization that combines features of FISH and polymerase chain reaction (PCR). It is specific, simple, cost effective and rapid. In contrast to FISH, it does not require a previously labeled probe. Instead, the labeling reaction takes place in situ. Koch et al. [1] initially described this technique and later was improved extensively by Pellestor et al. [2] and Tharapel et al. [3]
The demand for prenatal rapid aneuploidy diagnosis is increasing, as this is the only method for prevention of common aneuploidies. Presently aneuploidy diagnosis is time consuming, expensive and with significant failures in particular with prenatal samples. FISH is a good and rapid method; however, it is expensive and takes over 24 h. This is because there is no centromere specific FISH probe for chromosome 13 and 21. Centromeric probes provide good signal in very short time (~6 h).
This study evaluated the application of PRINS for rapid diagnosis of common chromosomal aneuploidies (X, Y, 13, 18, and 21). We used PRINS primers specific for chromosome X, Y, 13, 18, and 21 centromeric or heterochromatic repeat sequences, and modified the technique to allow greater hybridization specificity.
 Materials and Methods Materials and Methods | |  |
This study was performed in the Department of Reproductive Biology, AIIMS, New Delhi. PRINS technique is based on in situ annealing of a specific oligonucleotide primer, followed by primer elongation by Taq DNA polymerase in presence of labeled nucleotides viz., Fluorescein Isothiocyanate (FITC), rhodamine, Cyanine 3 (Cy3), etc.,). It starts with in situ hybridization (annealing) of synthetic oligonucleotide to denatured complementary nucleic acid sequence. The oligonucleotide forms hybrids with its complementary target sequence, and then acts as a primer for the synthesis of labeled DNA by extension from the 3' end at the target site. After a few minutes of extension of labeled DNA, the reaction is stopped and signals visualized by fluorescence microscopy.
Informed consent was obtained and peripheral blood samples were collected from 10 healthy men. Standard cytogenetic techniques were used to obtain lymphocytes metaphase chromosomes from short-term blood micro cultures. Clean and dry glass slides were used for slide preparation. Interphase and metaphase chromosome were freshly prepared and fixed in 3:1 methanol acetic acid after hypotonic treatment with 50 mM KCl for 20 min. Slides were dehydrated by passing through ethanol series (70%, 90%, and 100%) and then air-dried.
We used one primer each (synthesized from Sigma, India) specific for centromere/alphoid of chromosome X, Y, 13, 18, and 21 [[Table 1]; Choo et al. [4] ]. We prepared about 25-μl PRINS reaction mixture containing individual primer (for 5 primers five reaction mixture containing one primer each; vide [Table 1] for primer amount), Deoxyribonucleotide Triphosphate (dNTPs; 0.2 mM; N = Adenine/Cytosine/Guanine; Promega Corporation, USA), Deoxyribothymine Triphosphate (dTTP; 0.02 mM; Promega Corporation, USA), Cyanine 3-12-Deoxyribouracil Triphosphate (Cy3-12-dUTP; 0.02 mM; Amersham, UK), KCl (50 mM, Sigma, India), Tris-HCl (10 mM, pH 8.3, Sigma, India), MgCl 2 (2 mM, Sigma, India), Bovine Serum Albumin (BSA) (0.01%, Roche, Germany) and Taq DNA polymerase (1 Unit; Promega, USA). DNA denaturation of the metaphase spreads was carried out by incubating the slides in solution containing 70% deionized formamide and 2X SSC at 76°C for 6 min, followed by successive passes through ice-cold ethanol baths (70%, 90%, and 100%) for 2 min each, and then air-dried. The 25-μl PRINS reaction mixture was heated to 76°C for 6 min before adding to the denatured metaphase spread slide, covered with a coverslip and sealed with rubber cement (to prevent evaporation). The slide was then placed on a FISH hybrite and incubated at 55°C to 68°C (according to the primer sequence; vide [Table 1]) for 60 min to allow primers to anneal, and then at 72°C for 10 min to allow primer extension. The coverslip was removed, and slide was placed in 0.4X SSC, 0.3% NP-40 (nonyl phenoxypolyethoxylethanol, Sigma, India) at 72°C for 2 min followed by 2X SSC/0.1% NP-40 for another 2 min to remove unbound primers and unincorporated dNTPs. Slides were then dehydrated in ethanol series (70%, 90%, and 100%) for 2 min in each before air-drying in darkness. About 10 μl of 4', 6-diamidino-2-phenylindole (DAPI) solution (100 μg/ml) containing antifade-mounting medium was applied to the target area under a cover glass. Excess of antifade was removed and slides were sealed with nail varnish. Microscopy was done under fluorescent microscope (Olympus BX51, Olympus, Japan) and result was analyzed by two independent observer using appropriate filters. Photomicrography was taken with the help of FISH Imaging System Applied Spectral Imaging; CA, USA.
 Results Results | |  |
About 10 experiments were carried out with each primers, specific signals were visualized with chromosome X, Y, 13, 18, and 21 [Figure 1]a-e. PRINS results were satisfactory as signals were specific, discrete, uniform, repeatable and prominent with X, Y, 13, 18, and 21 primers.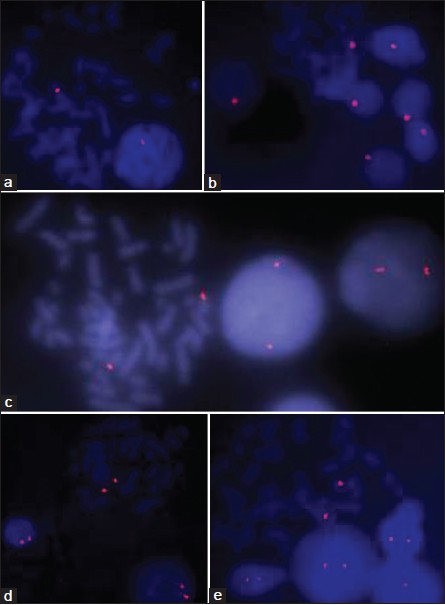 | Figure 1: Primed in situ labeling/synthesis images on metaphase as well as interphase cells with Chromosome X (a), Y (b), 13 (c), 18 (d), and 21 (e) specific primers
Click here to view |
 Discussion Discussion | |  |
Chromosomal abnormalities are a major cause of genetic disease. It accounts for a large proportion of reproductive wastage and congenital malformations. Cytogenetic analysis by conventional chromosomal banding techniques, although highly precise and an important standard method, requires cell culture, skilled personnel and is labor intensive. Conventional cytogenetic is time consuming, expensive and with significant failures in particular with prenatal samples. Conventional cytogenetics is possible only in dividing cells. These factors have led investigators to seek newer approaches for identifying chromosomal abnormalities. These newer approaches (molecular cytogenetics) can be applied throughout cell cycle, in non-dividing cells, dead cells, and fixed cells. High sensitivity, specificity, and speed have made newer molecular approaches a powerful tool in modern cytogenetics. FISH is a good and rapid method; however, it is expensive and takes over 24 h. PRINS is a molecular cytogenetic technique based on in situ annealing of a specific oligonucleotide primer, followed by primer elongation by Taq DNA polymerase in presence of labeled nucleotides (e.g., FITC, Cy3, etc.). It is a fast alternative to FISH and requires only a few handling steps. In contrast to FISH, a previously labeled probe is not required. Instead, the labeling reaction takes place in situ. The goal of this study was to standardize and evaluate whether PRINS can be used for rapid and inexpensive diagnosis of common aneuploidies. As the conventional method of diagnosis is unsatisfactory due to time taken for prenatal diagnosis and expense of FISH probes, this might have tremendous promise.
This work was designed to evaluate the application of PRINS for rapid diagnosis of common chromosomal aneuploidies (X, Y, 13, 18, and 21). Presently, chromosome 21 and 13 centromere specific FISH probes are unavailable. Centromeric probes provide big signals and diagnosis can be made in 6-8 h whereas locus specific probes takes more than 24 h. PRINS is a modified PCR based technique and any centromeres of chromosome can be amplified specifically, hence we tested this method for aneuploidy diagnosis to overcome difficulties of FISH. We found that method is working, repeatable and reliable. We conclude that PRINS is reliable at common aneuploidy diagnosis and may be used in diagnosis of common aneuploidies rapidly as well as cost-effective. Rapid prenatal diagnosis of common aneuploidies (13, 16, 18, 21, X, and Y) in interphase and metaphase nuclei by PRINS was previously described by Li et al. [5] Similarly, preconception screening of polar bodies for common aneuploidy (13, 16, 18, 21, or X) by PRINS method was described by Petit et al. [6] Our experience with centromeres/alphoid specific (X, Y, 13, 18, and 21) PRINS indicates that PRINS is a reliable technique for rapid aneuploidy diagnosis.
 Acknowledgment Acknowledgment | |  |
The study was supported by grants from Indian Council of Medical Research and AIIMS, New Delhi, India.
 References References | |  |
| 1. | Koch JE, Kølvraa S, Petersen KB, Gregersen N, Bolund L. Oligonucleotide-priming methods for the chromosome-specific labelling of alpha satellite DNA in situ. Chromosoma 1989;98:259-65. 
|
| 2. | Pellestor F, Andréo B, Coullin P. Interphasic analysis of aneuploidy in cancer cell lines using primed in situ labeling. Cancer Genet Cytogenet 1999;111:111-8. 
|
| 3. | Tharapel SA, Kadandale JS. Primed in situ labeling (PRINS) for evaluation of gene deletions in cancer. Am J Med Genet 2002;107:123-6. 
|
| 4. | Li F, Fang W, Wu B, Wang J, Zhong H, Heng W. Molecular cytogenetic detection of trisomy 21 in interphase nuclei and metaphase chromosomes. Chin Med J (Engl) 1999;112:840-4. 
|
| 5. | Petit C, Martel-Petit V, Fleurentin A, Monnier-Barbarino P, Jonveaux P, Gerard H. Use of PRINS for preconception screening of polar bodies for common aneuploidies. Prenat Diagn 2000;20:1067-71. 
|
| 6. | Choo KH, Vissel B, Nagy A, Earle E, Kalitsis P. A survey of the genomic distribution of alpha satellite DNA on all the human chromosomes, and derivation of a new consensus sequence. Nucleic Acids Res 1991;19:1179-82. 
|
[Figure 1]
[Table 1]
|






