|
 
 |
|
ORIGINAL ARTICLE |
|
|
|
| Year : 2013 | Volume
: 19
| Issue : 1 | Page : 32-42 |
| |
Rapid-prenatal diagnosis through fluorescence in situ hybridization for preventing aneuploidy related birth defects
Ashish Fauzdar1, Mohit Chowdhry1, RN Makroo1, Manoj Mishra1, Priyanka Srivastava1, Richa Tyagi1, Preeti Bhadauria1, Anita Kaul2
1 Department of Transplant Immunology, Molecular Biology and Transfusion Medicine, Sarita Vihar, New Delhi, India
2 Department of Fetal Medicine, Apollo Hospitals, Sarita Vihar, New Delhi, India
| Date of Web Publication | 4-Jun-2013 |
Correspondence Address:
Ashish Fauzdar
Department Transplant Immunology, Molecular Biology and Transfusion Medicine; Indraprastha Apollo Hospitals, New Delhi
India
 Source of Support: None, Conflict of Interest: None
DOI: 10.4103/0971-6866.112881

 Abstract Abstract | | |
Background and Objective: Women with high-risk pregnancies are offered prenatal diagnosis through amniocentesis for cytogenetic analysis of fetal cells. The aim of this study was to evaluate the effectiveness of the rapid fluorescence in situ hybridization (FISH) technique for detecting numerical aberrations of chromosomes 13, 21, 18, X and Y in high-risk pregnancies in an Indian scenario.
Materials and Methods: A total of 163 samples were received for a FISH and/or a full karyotype for prenatal diagnosis from high-risk pregnancies. In 116 samples both conventional culture techniques for getting karyotype through G-banding techniques were applied in conjunction to FISH test using the AneuVysion kit (Abbott Molecular, Inc.), following standard recommended protocol to compare the both the techniques in our setup.
Results: Out of 116 patients, we got 96 normal for the five major chromosome abnormality and seven patients were found to be abnormal (04 trisomy 21, 02 monosomy X, and 01 trisomy 13) and all the FISH results correlated with conventional cytogenetics. To summarize the results of total 163 patients for the major chromosomal abnormalities analyzed by both/or cytogenetics and FISH there were 140 (86%) normal, 9 (6%) cases were abnormal and another 4 (2.5%) cases were suspicious mosaic and 10 (6%) cases of culture failure. The diagnostic detection rate with FISH in 116 patients was 97.5%. There were no false-positive and false-negative autosomal or sex chromosomal results, within our established criteria for reporting FISH signals.
Conclusion: Rapid FISH is a reliable and prompt method for detecting numerical chromosomal aberrations and has now been implemented as a routine diagnostic procedure for detection of fetal aneuploidy in India.
Keywords: Aneuploidy, birth defects, fluorescence in situ hybridization, prenatal diagnosis
How to cite this article:
Fauzdar A, Chowdhry M, Makroo R N, Mishra M, Srivastava P, Tyagi R, Bhadauria P, Kaul A. Rapid-prenatal diagnosis through fluorescence in situ hybridization for preventing aneuploidy related birth defects. Indian J Hum Genet 2013;19:32-42 |
How to cite this URL:
Fauzdar A, Chowdhry M, Makroo R N, Mishra M, Srivastava P, Tyagi R, Bhadauria P, Kaul A. Rapid-prenatal diagnosis through fluorescence in situ hybridization for preventing aneuploidy related birth defects. Indian J Hum Genet [serial online] 2013 [cited 2016 May 24];19:32-42. Available from: http://www.ijhg.com/text.asp?2013/19/1/32/112881 |
 Introduction Introduction | |  |
Birth defects occur as a result of genetic problems caused by mutations in one or more genes, chromosomal aneuploidy or environmental factors during the gestational period. Cytogenetic disorders occur in about 2% of pregnancies in women older than 35 years, 1% of live births and 6% of still births. It is evident that more than 50% of first trimester spontaneous abortions are due to chromosomal abnormalities. [1],[2] In India, almost half a million babies are born annually with malformations; the figure for Down syndrome (trisomy 21) is 21,000 (21) or 1/1150 births. [3] This load is more common than any other genetic disorder. Moreover, in pregnancy with ultrasound detected malformations, its incidence is much higher and varies from 17% to 27%. [4],[5],[6] The most common cause of spontaneous abortion is numerical chromosome imbalances (aneuploidies), particularly of chromosomes 13, 14, 15, 16, 18, 21, 22, and X. [7],[8],[9] Aneuploidies of five particular chromosomes (13, 18, 21, X, Y) accounts for 95% of the chromosomal aberrations that lead to infants born with congenital defects. [10] Therefore, it is essential in prenatal diagnostics to analyze chromosomal abnormalities by utilizing procedures such as amniocentesis and chorionic villus sampling (CVS) for early detection of potential birth defects, particularly in high risk pregnancies.
The conventional full karyotype of amniocytes and cytotrophoblasts is labor intensive, time consuming and requires highly skilled personnel for accurate analysis. Internationally, the turnaround rate for numerical aberration detection, with respect to chromosomes 13, 18, 21, X, and Y, using rapid fluorescence in situ hybridization (FISH) on amniocytes, has tremendously decreased from few hours, 2 h to 48 h over a period of time. [11],[12],[13],[14],[15],[16],[17],[18] The FISH allows for the analysis of chromosome aberrations using chromosome specific probes. Currently, most laboratories perform rapid tests for aneuploidy such as FISH as part of a combination test with traditional cytogenetics for prenatal diagnosis in India.
Recent development of newer techniques have guaranteed a significantly shortened turnaround time for obtaining results in prenatal diagnosis. These modern techniques include the aforementioned FISH method, multi-primed in situ labeling (multi-PRINS), quantitative fluorescence polymerase chain reaction (QF-PCR) and multiplex ligation-dependent probe amplification (MLPA). [11],[19],[20],[21],[22] There have been few studies on the applications of molecular cytogenetics for prenatal diagnostics in India. [12],[23] Moreover, there have been no publications in this area following the printing of the aforementioned articles, as per the MEDLINE search till December, 2010. The aim of the present study was to evaluate the accuracy of the rapid FISH technique for the early detection of numerical aberrations of chromosomes 13, 21, 18, X, and Y in interphase nuclei of uncultured amniocytes/cytotrophoblast cells in 163 high-risk pregnancies in comparison with conventional cytogenetics.
 Materials and Methods Materials and Methods | |  |
Prenatal diagnosis was performed on 163 (162 singlet 01 twin) high-risk pregnancies. Samples were obtained from amniotic fluid (129), chorionic villus (31), cord blood (2) and fetal urine (1). A retrospective study was conducted on the tests conducted over the course of approximately 4 years, beginning from year 2006 to 2010, at the Department of Fetal Medicine and Immunology and Molecular Biology, Indraprastha Apollo Hospitals, New Delhi. The indications of high-risk pregnancies suggested for further prenatal testing for detection of numerical chromosome aberrations are depicted in [Table 1]. Samples for invasive prenatal diagnosis were taken only after proper genetic counseling and signing of the consent form (Pre-conception and Prenatal Diagnostic Techniques [Prohibition of Sex Selection] Amendment Act, 2002) by both patient and clinician.
The sample was taken either through CVS (at 11-13 weeks) or an amniocentesis (at 16-18 weeks) under ultrasound guidance by the fetal medicine department. After procedure 10-20 ml of amniotic fluid or 5-10 mg of chorionic villus was obtained from each patient depending on the gestation stage at the time of sampling. Amniotic fluid and chorionic villi samples were then subjected to FISH and full karyotype or only FISH, for chromosome analysis as per request of clinician. A rapid FISH analysis was performed on interphase cell preparation as described below (from amniotic fluid, chorionic villus, fetal urine, and cord blood sample). Altogether 116 samples were analyzed through Rapid FISH in combination with conventional cytogenetics successfully. Standard culture and harvesting methods for chromosome preparations along with G-banding were followed for amniotic fluid and chorionic villus sample. [24],[25] The FISH analysis was performed on uncultured amniocytes using DNA probes specific for chromosomes 13, 18, 21, X and Y using US Food & Drug Administration (FDA) cleared AneuVysion kit.
Preparation of uncultured cells for interphase cells for FISH
Amniocytes
Five milliliter to ten milliliter of amniotic fluid was centrifuged (1000 rpm, 5 min) and the pellet was suspended in 5 ml 1X Trypsin/ Ethylene Diammine Tetra Acetate (EDTA) (Biological Industries Israel, cat 03-051-58) and incubated for 30 min at 37°C. After centrifugation, the pellet was resuspended in 7 ml 0.075 M KCl and incubated at 37°C for 15 min. After addition of a few drops of Carnoy's fixative (methanol: Acetic acid [3:1]), the pellet was centrifuged, and resuspended in Carnoy's fixative for washing at least three times. Subsequently, the cells were incubated at −20°C for 20 min in 5 ml Carnoy's fixative and the preparation was completed with a final centrifugation.
Chorionic villous cells
Five to ten milligram (1-2 pieces) of villus are cleaned in Rosewell Park Memorial Institute (RPMI) medium in 35 mm petridish and are transferred to 15 ml centrifuged tube and incubated in 5 ml 1X trypsin/EDTA and kept at 37°C for 10 min. After incubation centrifugation was done at 1000 rpm for 10 min to remove the supernatant and further incubate in 0.075 M KCl (hypotonic solution) at 37°C for 40 min. After incubation we add few drops of Carnoy's fixative, then pellet was centrifuged, and resuspended in fixative for washing at least three times. Finally, the villi are dissociated by adding 60% acetic acid followed by continuous vortexing and mixing to get isolated cells.
Fetal urine cells
Urine sample was collected in 15 ml centrifuged tube and is centrifuged at 1000 rpm for 10 min. Supernatant was discarded and pellet was washed three times in 1X phosphate buffer saline (PBS). After washing, we add in 0.075 M KCl (hypotonic solution) and incubate at 37°C for 30 min. After incubation, we add few drops of fixative, followed by centrifugation and resuspension in fixative. Cells are then washed further at least for three times to get clean white pellet of cells.
Umbilical cord blood
One milliliter to two milliliter of cord blood sample was collected in 15 ml centrifuged tube and washed to remove debris by adding 1X PBS which is centrifuged at 1000 rpm for 10 min. After washing to the sample, we add in 0.075 M KCl (hypotonic solution) and incubated at 37°C for 30 min. After incubation, we add few drops of fixative, then centrifuged to get pellet, finally resuspended in fixative followed by washing at least for 3 times.
Rapid fluorescence in situ hybridization
Rapid fluorescence in situ hybridization (FISH) was performed on uncultured amniocytes/CVS sample using DNA probes specific for chromosomes 13, 18, 21, X and Y following modified standardized protocol through rapid aneuploidy screening by AneuVysion kit (Abbott/Vysis; Downers Grove, IL). [26] After interphase cell preparation, cells were resuspended in 200 μl of Carnoy's fixative (methanol: Acetic acid [3:1]) and dropped on a clean cold slides kept at 4°C. We drop about ~50 μl of cell suspension on each of the two separate hybridization areas marked with the glass marker. Slides were then taken for FISH procedure, first the slides are dipped for 10 min at 37°C the slides in 2X SSC [3 M sodium chloride (Fisher Scientific cat. No 27605) and 300 mM tri-sodium citrate 2-hydrate (Merck cat. No 17556)] then slides are dipped for 10 min at 37°C into protease solution (43 μl 12N HCl, 25 mg Pepsin [Sigma-Aldrich cat. No P6887] in 50 ml milliQ water) followed by in 1X PBS for 5 min. Then slides are dipped in neutral buffer formalin buffer (180 mg Mgcl 2 .6H 2 0 [Amresco cat. No 0288-100G], 1 ml 37-41% w/v formaldehyde [Merck cat. No. 61780850001730] in 39 ml 1X PBS) for 5 min at room temperature (RT) followed by in 1X PBS for 5 min and then are dehydrated in series of 70%, 85% and 100% ethanol for 5 min each. Finally, slides are air dried before addition of probe 13, 21 and X, Y, 18 in both the hybridization area followed by co-denaturation of slide and probe at 90°C for 5 min. The slides are then kept for hybridization at 37°C for overnight incubation in air tight humidified box. Next day, the post-hybridization washings was carried out in 0.4X SSC/03% NP-40 at 73°C for 2 min followed by 2X SSC/0.1% NP-40 for 30 sec. Subsequently slides are air dried and counterstained with 8 μl 4, 6-diamidino-2-phenylindole (DAPI) containing anti-fade and a cover slip. The slides are then incubated at −20°C for at least 10 min prior to signal enumeration under the fluorescence microscope. The slides were screened for 50 interphase nuclei per case and probe combination. The signal analysis was carried out using an Olympus fluorescent microscope model BX60 equipped with a 100-watt mercury bulb, 100X plane apochromatic objective and single band pass filter for DAPI, fluorescein-5-and/or-6 isothiocyanate (FITC) and tetramethyl rhodamine isothiocyanate (TRITC) and a dual band pass filter for TRITC and FITC (Olympus Japan). Image acquisition was performed with an epifluorescence microscope (Olympus BX 60) with a cooled charge-coupled device (CCD) camera with karyotype software package (Cytovision, Applied Imaging, Sunderland, UK). Turn around time for getting FISH results in our cases was 48-72 h from time of receipt of the sample.
Probes
We exclusively utilized an FDA-approved FISH test kit for rapid aneuploidy screening in uncultured amniocytes and chorionic villus sample; the AneuVysion kit (commercially available from Abbott/Vysis; Downers Grove, IL) consisted of three satellite DNA probes for Chromosomes Enumeration Probes X, Y, and 18 (CEP X, CEP Y, and CEP 18) and two locus-specific Identifier probes for 13q14 (LSI13) and 21q22.13 ~ 22.2 (LSI 21). The three centromeric probes and the two locus-specific probes were applied to the samples in two separate hybridizations on the glass slide.
Signal interpretation
Results were enumerated on the counting of 50 interphase nuclei per target and are reported as the number and percentage of nuclei with 1, 2, 3, 4, and >4 signals for CEP 18, LSI 13, and LSI 21 and as the number and percentage of nuclei with X, Y, XX, XY, XXY, XYY, XXX, and other, for CEP X/Y. The criteria for reporting the cutoff level by for FISH data is adopted from the previous established criteria. [26] A case was classified as informative normal for a specific chromosome if in more than 90% of the nuclei from each chromosomal (autosome/sex chromosome) hybridization demonstrated two normal disomic signal pattern. A case was considered as informative abnormal if more than 70% of the nuclei from each chromosome (autosome/sex chromosome) showed 3 signals or aberrant signal pattern. In cases where we observe 10-60% aberrant signal pattern were taken as suspicious mosaic and were considered as uninformative through FISH.
Conventional cytogenetics for amniocytes/chorionic villus culture
Amniotic fluid culture
Five milliliter to ten milliliter of amniotic fluid was centrifuged and supernatant was discarded, pellet was resuspended in 1 ml of Gibco® Amniomax TM -II Complete medium (cat no.11269-016). Cells are then seeded in T25 culture flask with additional 5 ml medium kept at 37°C with 5% CO 2 and growth of the cells was monitored for next 6-7 days. After the flask become confluent with the cell growth we add 100 μl Colcemid solution, 10μg/ml (Biological Industries, cat. No. 12-004-ID) and further incubated for 60 min at 37°C. After incubation 5 ml of 1X Trypsin-EDTA was added at RT for 5 min for cell dissociation. Entire content including cells are transferred in the centrifuge tube and centrifuged at 1000 rpm for 10 min, supernatant is discarded pellet is further resuspended in 7 ml of hypotonic solution (0.075 M KCl + 0.0052 M tri-sodium citrate in 1:1 ratio). The cells are incubated at 37°C for 25 min and cell and are fixed with few drops of fixative after incubation and further washing is carried out with carnoys fixative until we get the white pellet of cells. Then cell pellet is dropped on cold clean slides and aged for overnight at 65°C and then taken for banding with standard protocol through trypsin-geimsa banding. [27] Image acquisition was performed with an epifluorescence microscope (Olympus BX 60) with a cooled CCD camera with karyotype software package (Cytovision, Applied Imaging, Sunderland, UK) and 20 metaphase were captured for karyotype analysis. Turn around time for getting karyotype through conventional cytogenetics in amniotic fluid sample was 3-4 weeks from time of receipt of the sample depending on the growth of amniocytes.
Chorionic villus culture
Five milligram to ten milligram (1-2 pieces) of villus is thoroughly cleaned in RPMI medium in 35 mm petridish to remove the maternal decidua to avoid the maternal cell contamination. Cleaned villi are transferred in a centrifuge tube and pre-warmed (37°C) collagenase (solution type III [Sigma cat. No. C0255], 500 u/ml) is added and the incubated at 37°C water bath for 30-40 min until the supernatant become granular (leading to small pieces of cytotrophoblast). After complete digestion of the villi tube is centrifuged and supernatant is discarded, pellet is then resuspended in 1 ml of changes medium (Irvine Scientific cat. No. C100). Cells are then seeded in T25 culture flask with additional 5 ml medium and kept at 37°C with 5% CO 2 and growth of the cells is monitored for next 6-7 days till the flask become confluent with the cell growth. The harvesting and fixation of the cells was carried out as per the standard protocol as described above for amniotic fluid culture. After harvesting cell pellet is dropped on clean cold slides and taken for banding with standard protocol through trypsin-geimsa banding. Slides are taken for capturing and analyzing karyotype analysis for 20 metaphases at least. Turnaround time for getting metaphase preparation and subsequently getting results through conventional cytogenetics in chorionic villus sample was 3-4 weeks.
 Results Results | |  |
Of the total, 163 patients who underwent prenatal diagnosis, the median maternal age was 32 years (SD ± 5.3) (range 18-45); the median gestational age was 18 weeks (SD ± 6.9) (range 14-24 weeks). Detection of chromosomal abnormalities in high-risk pregnancies was accomplished via both/or FISH, conventional cytogenetics. In our study, the indications for prenatal diagnosis includes, positive triple test n0 = 49 (30.0%), abnormal ultrasonography (USG) n = 44 (27.0%), advanced maternal age + abnormal triple test n0 = 23 (14.0%), only advanced maternal age n = 11 (7.0%), triple test + abnormal USG n0 = 08 (5.0%), first trimester screening n = 07 (5.0%), β-thalassemia carrier parents n = 03 (2.0%), chromosome abnormality in family n = 04 (2.0%) and other's n = 14 (8.0%) [Table 1].
Of the total 163 samples received, 116 patients requested both cytogenetic and FISH analysis of fetal cells (amniocytes/chorionic villus and cord blood). The samples were taken from amniotic fluid (90), chorionic villus (24), and umbilical cord blood (2) processed for cytogenetics and FISH technique as per the standard procedure described above. Successful results were obtained in 106 patients by both cytogenetics and FISH where we observed 96 (82.7%) informative normal for five major chromosome abnormalities (13, 21, 18, X, and Y) and 7 (6%) informative abnormal including trisomy 21 in four patients [Figure 1], monosomy X in two patients [Figure 2] and trisomy 13 in one patient [Figure 3]. All the normal and abnormal results obtained through FISH were later confirmed and correlated with cytogenetic studies. In three patients, we could get the structural abnormalities through conventional cytogenetics we got 46, XX inv (9) (p11q12), 47, XX inv (9) (p11q12) +13, and 46, XX 15ps +, all results including trisomy 13, which could be picked up in FISH except structural rearrangements. As per our reporting criteria for FISH signal enumeration of the 116 patients taken for both FISH and cytogenetics, three patients were suspicious mosaic giving aberrant FISH signals between 10% and 60% of nuclei with three spectrum orange signals resulting in trisomy for chromosome 21. These cases were reported as uninformative through FISH within our established criteria after final FISH signal analysis after analyzing more number of nuclei. These Conventional cytogenetics results in three suspected mosaic cases turned out to have normal karyotype after analyzing more than 20 metaphases. Details of all the patients with chromosome abnormalities detected through conventional cytogenetics and FISH are shown in [Table 2].
In ten patients, cytogenetic analyses was incomplete due to culture failure of fetal cells; seven failures came from chorionic villus samples; two came from umbilical cord blood; one from amniotic fluid. We did FISH in all the ten cases of failed cytogenetics and no aneuploidy for the five chromosomes 13, 18, 21, X, and Y was observed in all cases. The diagnostic detection rate for only five chromosome analyzed through FISH in 116 patients was 97.5% as we got 03 uninformative cases, which were suspected mosaic and diagnostic detection rate with conventional cytogenetics was 91.4% due to 10 cases of culture failures. In 116 cases analyzed for both cytogenetic and FISH had an aneuploidy detection rate of 100% for identifying chromosomal abnormalities with 100% signal hybridization efficiency of probe in all the attempted cases. Out of total 163 samples only FISH test was carried out in 47 patients with normal disomic signal for chromosomes in 44 (94%) patients and 2 (5%) abnormal cases of trisomy 21 and 1 (1%) case was of suspected mosaic. To summarize all the results obtained out of total 163 patients through both/or cytogenetics and FISH, there were 140 (92%) patients normal for at least five common chromosome abnormality and 09 (5.5%) abnormal cases (06 trisomy 21, 02 monosomy X, and 01 trisomy 13) and 04 (2.5%) cases were suspicious mosaic through FISH [Table 3].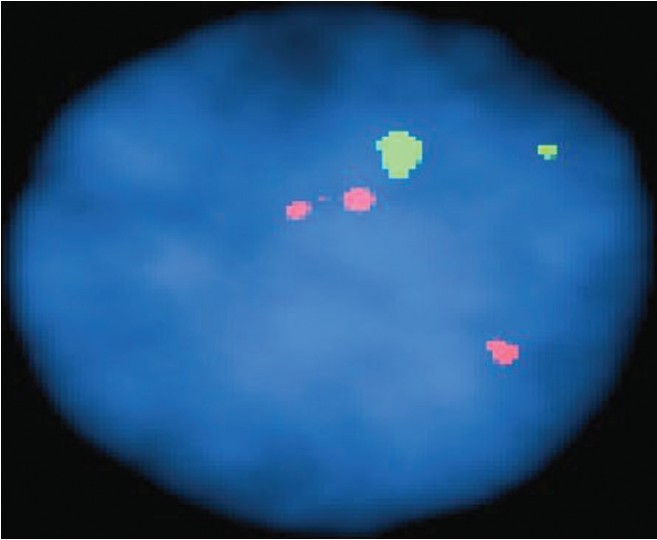 | Figure 1: Patient P1 with cytogenetics 47, **+21 showing trisomy 21 with three spectrum orange signals for chromosome 21 and two spectrum green signal for chromosome 13 after doing fluorescence in situ hybridization in amniocytes nuclei
Click here to view |
 | Figure 2: Patient P2 with cytogenetics karyotype showing 45* showing only one spectrum green signal for chromosome X after doing fluorescence in situ hybridization in amniocytes nuclei
Click here to view |
 | Figure 3: Patient P5 with representative cytogenetics karyotype 47 **inv (9) (p11q12) +13 showing trisomy 13 with three spectrum green signals for chromosome 13 and two spectrum orange signal for chromosome 21
Click here to view |
 | Table 2: Details of patients with chromosome abnormalities through conventional cytogenetics and FISH
Click here to view |
 Discussion Discussion | |  |
Our study was designed to aid both the patient and clinician in management of the high risk pregnancies by providing timely, accurate results on chromosomal abnormalities via FISH as well as conventional karyotyping. Rapid and accurate detection of chromosomal aneuploidies has been demonstrated by several researchers which compared aneuploidy detection by interphase FISH for chromosome 13, 18, 21, X, and Y with conventional cytogenetics. [11],[15],[28],[29],[30],[31] There have been few reports in the area of prenatal diagnostics for chromosomal disorders from India. [13],[29],[32] After the publication of the previously mentioned articles, few studies have been reported even after several years in which molecular cytogenetic techniques such as FISH have been utilized on Indian patients for prenatal diagnosis.
In this study, we did FISH in 163 patients and got informative results in all the patients through both/or cytogenetics and FISH. In our study group 85% of cases were normal for at least five common chromosome abnormalities and 6.0% were abnormal and 2.5% cases were considered to be suspicious mosaic through FISH. We got aneuploidy rate of 6.0% and diagnostic detection rate of 97.5% by FISH due to four uninformative cases. 116 cases analyzed by both cytogenetic and FISH had an aneuploidy detection rate of 100% as we got results for all the five major chromosome abnormalities tested in all cases including in the failed conventional cytogenetics. Accurate determination of sensitivity, specificity and positive and negative value was not possible due to lack of follow-up data with postnatal karyotype and due to small numbers of cases for identifying chromosome aberrations by both FISH and routine cytogenetics. There were no false-positive or false-negative for autosomal or sex chromosomal results within our established criteria of reporting FISH signals.
Among the 116 patients taken for both FISH and cytogenetic three cases were considered suspicious mosaic giving aberrant FISH signals between 10% and 60% of nuclei within our established reporting criteria as described above. In these cases additional 200 nuclei were counted to confirm the signals and routine cytogenetics analysis was performed on larger number of metaphases. All the three cases were reported as uninformative through FISH after final signal enumeration. After conventional cytogenetics results in these three suspected mosaic cases turned out to have normal karyotype after analyzing more than 20 metaphases and no mosaic cell line were observed. There was one additional case where fetal urine cells were obtained and only FISH was carried out resulting in 15% aberrant signal pattern and cytogenetic analysis was not possible due to inappropriate sample for routine cytogenetics analysis [Table 3].
The American College of Medical Genetics (ACMG, 2000) and the American Society of Human Genetics (ASHG) have published guidelines endorsing the use of FISH tests for prenatal testing. [33] Whenever 10-60% cells express the same signal pattern suggestive or aneuploidy suspect's mosaicism and are considered uninformative through FISH. In these cases, FISH study is recommended by analyzing additional 100-200 nuclei with extended cytogenetic analysis. In these cases of suspicious mosaic through FISH with aberrant signals, it is wise to report them as inconclusive or uninformative test and waits for confirmation by standard karyotyping. ACMG also states that prenatal FISH tests provide highly accurate results for select chromosomal abnormalities and FISH results can be reported to the physician before the conventional chromosome analysis results are available. Similar to other areas of diagnostic testing, it recommends that irreversible decisions to act on positive results should be supported by two of the three possible pieces of information, i.e., FISH, conventional cytogenetics and clinical information. [34] In our four patients, proper genetic counseling was carried out by clinician and was discussed with the available options to the patient. After counseling three patients decided to terminate the pregnancy in lieu of the indications for prenatal diagnosis and one patient decided to continue the pregnancy and gave a live birth, postnatal karyotype results was not done till the compilation of the results.
In three cases, we got the structural abnormalities through conventional cytogenetics method, which could not be identified through FISH as the technique has limitation in identifying the structural aberrations and chromosome abnormalities other than five chromosomes included in the panel. We got chromosome nine inversion (structural aberration) in two cases through karyotype. However, in the second case we could detect trisomy 13 with three spectrum green signal through FISH. In the third case we obtained increase in the length of the satellite on the short arm of chromosome 15 and all the results in these cases correlated with the FISH study. Interphase FISH analysis for prenatal detection of the common aneuploidies for chromosome 13, 18, 21, X, and Y is accurate and reliable test. However, patient is thoroughly counseled by the clinician what this specific test can do and cannot do. Aneuploidies of chromosome other than 13, 18, 21, X, and Y, structural aberrations, ring or marker chromosomes and many mosaic states are theoretically undetectable by routine interphase FISH testing and therefore, it does not substitute for complete standard cytogenetics. Interphase FISH for detection of the common aneuploidies misses about 30% of all chromosome abnormalities detectable by standard cytogenetics even with 100% accuracy of the test. [35],[36] It is evident that considerable percentage of "missed" cases would include some de novo clinically significant abnormalities like balanced translocation or marker chromosomes. Hence, one of the major limitations of interphase FISH analysis is to provide information regarding only the specific probe loci used. The test allows counting of loci and has no power to detect rearrangements. So the patients are counseled for doing classical cytogenetics in conjunction to FISH for identification of rearrangements to reveal the mechanism of aneuploidy (translocation vs. trisomy) for planning future pregnancies.
Through conventional cytogenetics we could get informative results in 91.4% cases as there were ten cases of culture failures resulting in 8.6% of culture failure rate (10 cases out of 116), which is very high as compared to the previous studies. Out of 10 culture failures in the study there were seven CVS samples, two amniotic fluid and one cord blood samples. Culture failure rate in only amniotic fluid samples was 1% comparable to the other world-wide studies, which reported a culture failure rate of <1%. [37],[38] The culture failure problem was mainly in CVS samples included in initial part of standardization process. However, we could assist these patients by getting information about at least five chromosomes through FISH, which were found to be normal and subsequently patient was counseled for failed culture report by the clinician in lieu of indications for prenatal diagnosis. Conventional cytogenetics is currently standard prenatal diagnostic test and is routinely offered to patients having increased risk of carrying chromosomally abnormal fetuses. The traditional "gold standard" for prenatal diagnosis of chromosome abnormalities is metaphase analysis through G banding. The primary advantages of standard cytogenetic analysis are the ability to detect aneuploidies as well as structural chromosomal aberration with great accuracy. Karyotyping requires isolation of metaphase chromosomes from cultured fetal cells and therefore is time consuming. Though, the reporting time has decreased considerable in last few decades, conventional karyotyping still requires 7-14 days of which culture is the most time consuming. [39] However, cultures required for karyotyping can, at times, fail to grow sufficiently. Furthermore, karyotyping requires great technical expertise and time for chromosome organization. The lengthy turnaround time for the conventional method is not acceptable to many parents and obstetricians especially, during the second half of pregnancies because, in many countries, the legal limit of pregnancy termination is 20-22 weeks.
Hence, there is a need for a speedy alternative method and these factors have led investigators to seek other methods for identifying chromosomal abnormalities quickly through alternative techniques like rapid FISH. FISH involves hybridization of fluorescently labeled specific probes to the patient's chromosomal DNA and followed by signal detection using a fluorescent microscope. FISH can be applied in both interphase and metaphase cells and therefore, does not require cultures cells for diagnosis and it is a very useful technique for rapidly determining the number of chromosomes in interphase cell using chromosome specific probes. [40],[41],[42],[43] Aneuploidies of chromosomes X, Y, 13, 18, and 21 and account for about 65% of all chromosomal abnormalities and encompasses approximately 95% of chromosome abnormality cases, which accompany birth defects in newborns. [44],[45],[46] With the introduction of multicolor, commercially available, highly specific and reliable probes significantly enhanced the overall performance of the test with quality control reagents as well as techniques in the development of standardized protocol leading to quality assurance in reporting the results. In our study, we exclusively used FDA cleared AneuVysion assay kit (Vysis, Inc.) to enumerate chromosomes 13, 18, 21, X, and Y in amniocytes and chorionic villus cells.
Early receipt of normal disomic results through rapid FISH has a positive effect on the mother by reducing anxiety as it's a reliably fast method for detecting numerical chromosomal aberrations in prenatal diagnosis and now subsequently been implemented as a routine diagnostic procedure in high-risk pregnancies for fetal aneuploidy in India. FISH using probes specific for chromosome 13, 18, 21, X, and Y has the potential to obtain results quickly and thus are capable of reducing parental anxiety and guiding further obstetric management. One may argue that cost benefit issues will direct towards abandoning the expensive cytogenetic analysis in favor of the faster, less expensive FISH technique. Many feel that the cost of the missed cases far outweighs the savings. [35]
 Conclusion Conclusion | |  |
Rapid FISH with AneuVysion probes (13, 18, 21, X and Y) is a preliminary test; it is often used in conjunction with full karyotype analysis The present study using FISH probes specific for chromosome 13, 18, 21, X and Y has the potential to find answers quickly (48-72 h) and is capable of reducing parental anxiety and of guiding further with better obstetric management in India. Although FISH is used as a preliminary test, it reduces the anxiety that parents feel during a pregnancy. It's comparatively low sensitivity, due to its limitations as discussed in identifying only the most common aneuploidies and structural rearrangements, makes this analysis helpful only in conjunction with conventional cytogenetics. We hope in coming year in India, we could move forward to new molecular cytogentics methods like QF-PCR, MLPA and prenatal chips using array-Comparative Genomic Hybridization (CGH), which definitely offers a number of advantages over conventional cytogenetic analysis and FISH. However, the increasing cost of prenatal diagnosis associated with newer molecular techniques will be a limiting factor for better management of high-risk pregnancies in India.
 References References | |  |
| 1. | Hassold TJ. A cytogenetic study of repeated spontaneous abortions. Am J Hum Genet 1980;32:723-30. 
|
| 2. | Jacobs PA. The chromosome complement of human gametes. Oxf Rev Reprod Biol 1992;14:47-72. 
|
| 3. | Verma IC. Challenges in human genetics in India in the new millennium. Indian J Pediatr 2000;67:809-11. 
|
| 4. | Rizzo N, Pittalis MC, Pilu G, Orsini LF, Perolo A, Bovicelli L. Prenatal karyotyping in malformed fetuses. Prenat Diagn 1990;10:17-23. 
|
| 5. | Wilson RD, Chitayat D, McGillivray BC. Fetal ultrasound abnormalities: Correlation with fetal karyotype, autopsy findings, and postnatal outcome - Five-year prospective study. Am J Med Genet 1992;44:586-90. 
|
| 6. | Nicolaides KH, Rodeck CH, Gosden CM. Rapid karyotyping in non-lethal fetal malformations. Lancet 1986;1:283-7. 
|
| 7. | Chandley AC. Infertility and chromosome abnormality. Oxf Rev Reprod Biol 1984;6:1-46. 
|
| 8. | Zenzes MT, Casper RF. Cytogenetics of human oocytes, zygotes, and embryos after in vitro fertilization. Hum Genet 1992;88:367-75. 
|
| 9. | Jacobs PA, Hassold TJ. The origin of numerical chromosome abnormalities. Adv Genet 1995;33:101-33. 
|
| 10. | Whiteman DAH, Klinger K. Efficiency of rapid in situ hybridization methods for prenatal diagnosis of chromosome abnormalities causing birth defects. Am J Hum Genet 1991;49:A1279. 
|
| 11. | Klinger K, Landes G, Shook D, Harvey R, Lopez L, Locke P, et al. Rapid detection of chromosome aneuploidies in uncultured amniocytes by using fluorescence in situ hybridization (FISH). Am J Hum Genet 1992;51:55-65. 
|
| 12. | Jobanputra V, Roy KK, Kucheria K. Prenatal detection of aneuploidies using fluorescence in situ hybridization: A preliminary experience in an Indian set up. J Biosci 2002;27:155-63. 
|
| 13. | Bryndorf T, Christensen B, Vad M, Parner J, Carelli MP, Ward BE, et al. Prenatal detection of chromosome aneuploidies in uncultured chorionic villus samples by FISH. Am J Hum Genet 1996;59:918-26. 
|
| 14. | Stephenson MD, Awartani KA, Robinson WP. Cytogenetic analysis of miscarriages from couples with recurrent miscarriage: A case-control study. Hum Reprod 2002;17:446-51. 
|
| 15. | Luquet I, Mugneret F, Athis PD, Nadal N, Favre B, Abel C, et al. French multi-centric study of 2000 amniotic fluid interphase FISH analyses from high-risk pregnancies and review of the literature. Ann Genet 2002;45:77-88. 
|
| 16. | Caine A, Maltby AE, Parkin CA, Waters JJ, Crolla JA, UK Association of Clinical Cytogeneticists (ACC). Prenatal detection of Down's syndrome by rapid aneuploidy testing for chromosomes 13, 18, and 21 by FISH or PCR without a full karyotype: A cytogenetic risk assessment. Lancet 2005;366:123-8. 
|
| 17. | Choolani M, Ho SS, Razvi K, Ponnusamy S, Baig S, Fisk NM, et al. FastFISH: Technique for ultrarapid fluorescence in situ hybridization on uncultured amniocytes yielding results within 2 h of amniocentesis. Mol Hum Reprod 2007;13:355-9. 
|
| 18. | Ho SS, Choolani MA. Flash FISH: Same day prenatal diagnosis of common chromosomal aneuploidies. Methods Mol Biol 2010;659:261-8. 
|
| 19. | Pertl B, Yau SC, Sherlock J, Davies AF, Mathew CG, Adinolfi M. Rapid molecular method for prenatal detection of Down's syndrome. Lancet 1994;343:1197-8. 
|
| 20. | Hochstenbach R, Meijer J, van de Brug J, Vossebeld-Hoff I, Jansen R, van der Luijt RB, et al. Rapid detection of chromosomal aneuploidies in uncultured amniocytes by multiplex ligation-dependent probe amplification (MLPA). Prenat Diagn 2005;25:1032-9. 
|
| 21. | Gadji M, Krabchi K, Drouin R. Simultaneous identification of chromosomes 18, X and Y in uncultured amniocytes by using multi-primed in situ labelling technique. Clin Genet 2005;68:15-22. 
|
| 22. | Van Opstal D, Boter M, de Jong D, van den Berg C, Brüggenwirth HT, Wildschut HI, et al. Rapid aneuploidy detection with multiplex ligation-dependent probe amplification: A prospective study of 4000 amniotic fluid samples. Eur J Hum Genet 2009;17:112-21. 
|
| 23. | Verma IC, Saxena R, Lall M, Bijarnia S, Sharma R. Genetic counseling and prenatal diagnosis in India - Experience at Sir Ganga Ram Hospital. Indian J Pediatr 2003;70:293-7. 
|
| 24. | Rooney DE, Czepulkowski BH. In: Rooney DE, Czepulkowski BH editors. Human Cytogentics A Practical Approach. Vol. 1, Vol. 2. New York: Oxford University Press; 1992. 
|
| 25. | Eiben B, Goebel R, Hansen S, Hammans W. Early amniocentesis - A cytogenetic evaluation of over 1500 cases. Prenat Diagn 1994;14:497-501. 
|
| 26. | Eiben B, Trawicki W, Hammans W, Goebel R, Epplen JT. A prospective comparative study on fluorescence in situ hybridization (FISH) of uncultured amniocytes and standard karyotype analysis. Prenat Diagn 1998;18:901-6. 
|
| 27. | Seabright M. A rapid banding technique for human chromosomes. Lancet 1971;2:971-2. 
|
| 28. | Witters I, Devriendt K, Legius E, Matthijs G, Van Schoubroeck D, Van Assche FA, et al. Rapid prenatal diagnosis of trisomy 21 in 5049 consecutive uncultured amniotic fluid samples by fluorescence in situ hybridisation (FISH). Prenat Diagn 2002;22:29-33. 
|
| 29. | Jobanputra V, Roy KK, Kriplani A, Kucheria K. Prenatal diagnosis of chromosomal abnormalities in women with high risk pregnancies. Indian J Med Res 2001;114:148-55. 
|
| 30. | Ward BE, Gersen SL, Carelli MP, McGuire NM, Dackowski WR, Weinstein M, et al. Rapid prenatal diagnosis of chromosomal aneuploidies by fluorescence in situ hybridization: Clinical experience with 4,500 specimens. Am J Hum Genet 1993;52:854-65. 
|
| 31. | Morris A, Boyd E, Dhanjal S, Lowther GW, Aitken DA, Young J, et al. Two years' prospective experience using fluorescence in situ hybridization on uncultured amniotic fluid cells for rapid prenatal diagnosis of common chromosomal aneuploidies. Prenat Diagn 1999;19:546-51. 
|
| 32. | Verma IC, Mathew S, Elango R, Khanna I. Prenatal diagnosis of chromosomal disorders in Delhi. Indian Pediatr 1990;27:459-62. 
|
| 33. | Test and Technology Transfer Committee, American College of Medical Genetics, 9650 Rockville Pike, Bethesda, MD 20814-3998, United States. Technical and clinical assessment of fluorescence in situ hybridization: An ACMG/ASHG position statement. I. Technical considerations. Test and Technology Transfer Committee. Genet Med 2000;2:356-61. 
|
| 34. | American College of Medical Genetics. Prenatal interphase fluorescence in situ Hybridization (FISH) policy statement. Am J Hum Genet 1993;53:526-7. 
|
| 35. | Evans MI, Henry GP, Miller WA, Bui TH, Snidjers RJ, Wapner RJ, et al. International, collaborative assessment of 146,000 prenatal karyotypes: Expected limitations if only chromosome-specific probes and fluorescent in-situ hybridization are used. Hum Reprod 1999;14:1213-6. 
|
| 36. | Lewin P, Kleinfinger P, Bazin A, Mossafa H, Szpiro-Tapia S. Defining the efficiency of fluorescence in situ hybridization on uncultured amniocytes on a retrospective cohort of 27407 prenatal diagnoses. Prenat Diagn 2000;20:1-6. 
|
| 37. | Zahed L, al-Oreibi G, Darwiche N, el-Khechen S. Karyotype of amniotic fluid cells at the AUB-MC results on 2000 cases. J Med Liban 2000;48:121-6. 
|
| 38. | Bell JA, Pearn JH, Wilson BH, Ansford AJ. Prenatal cytogenetic diagnosis - A current audit. A review of 2000 cases of prenatal cytogenetic diagnoses after amniocentesis, and comparisons with early experience. Med J Aust 1987;146:12-5. 
|
| 39. | Evans MI, Klinger KW, Isada NB, Shook D, Holzgreve W, McGuire N, et al. Rapid prenatal diagnosis by fluorescent in situ hybridization of chorionic villi: An adjunct to long-term culture and karyotype. Am J Obstet Gynecol 1992;167:1522-5. 
|
| 40. | Pinkel D, Straume T, Gray JW. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci U S A 1986;83:2934-8. 
|
| 41. | Pinkel D, Landegent J, Collins C, Fuscoe J, Segraves R, Lucas J, et al. Fluorescence in situ hybridization with human chromosome-specific libraries: Detection of trisomy 21 and translocations of chromosome 4. Proc Natl Acad Sci U S A 1988;85:9138-42. 
|
| 42. | Lichter P, Cremer T, Tang CJ, Watkins PC, Manuelidis L, Ward DC. Rapid detection of human chromosome 21 aberrations by in situ hybridization. Proc Natl Acad Sci USA 1988;85:9664-8. 
|
| 43. | Cremer T, Lichter P, Borden J, Ward DC, Manuelidis L. Detection of chromosome aberrations in metaphase and interphase tumor cells by in situ hybridization using chromosome-specific library probes. Hum Genet 1988;80:235-46. 
|
| 44. | Rhoads GG, Jackson LG, Schlesselman SE, de la Cruz FF, Desnick RJ, Golbus MS, et al. The safety and efficacy of chorionic villus sampling for early prenatal diagnosis of cytogenetic abnormalities. N Engl J Med 1989;320:609-17. 
|
| 45. | Lebo RV, Flandermeyer RR, Diukman R, Lynch ED, Lepercq JA, Golbus MS. Prenatal diagnosis with repetitive in situ hybridization probes. Am J Med Genet 1992;43:848-54. 
|
| 46. | Jacobs PA, Melville M, Ratcliffe S, Keay AJ, Syme J. A cytogenetic survey of 11,680 newborn infants. Ann Hum Genet 1974;37:359-76. 
|
[Figure 1], [Figure 2], [Figure 3]
[Table 1], [Table 2], [Table 3]
| This article has been cited by | | 1 |
Prenatal diagnosis of common fetal aneuploidies: Scenario in India |
|
| Baburao, V. and Gorakshakar, A.C. | | Indian Journal of Human Genetics. 2013; 19(1): 1-2 | | [Pubmed] | |
|
 |
|






