|
 
 |
|
ORIGINAL ARTICLE |
|
|
|
| Year : 2013 | Volume
: 19
| Issue : 3 | Page : 320-324 |
| |
No CAG repeat expansion of polymerase gamma is associated with male infertility in Tamil Nadu, South India
J Poongothai
Department of Biotechnology, PSG College of Technology, Coimbatore, Tamil Nadu, India
| Date of Web Publication | 30-Oct-2013 |
Correspondence Address:
J Poongothai
Department of Biotechnology, PSG College of Technology, Coimbatore - 641 004, Tamil Nadu
India
 Source of Support: None, Conflict of Interest: None
DOI: 10.4103/0971-6866.120823

 Abstract Abstract | | |
Mitochondria contains a single deoxyribonucleic acid (DNA) polymerase, polymerase gamma (POLG) mapped to long arm of chromosome 15 (15q25), responsible for replication and repair of mitochondrial DNA. Exon 1 of the human POLG contains CAG trinucleotide repeat, which codes for polyglutamate. Ten copies of CAG repeat were found to be uniformly high (0.88) in different ethnic groups and considered as the common allele, whereas the mutant alleles (not -10/not -10 CAG repeats) were found to be associated with oligospermia/oligoasthenospermia in male infertility. Recent data suggested the implication of POLG CAG repeat expansion in infertility, but are debated. The aim of our study was to explore whether the not -10/not -10 variant is associated with spermato g enic failure. As few study on Indian population have been conducted so far to support this view, we investigated the distribution of the POLG CAG repeats in 61 infertile men and 60 normozoospermic control Indian men of Tamil Nadu, from the same ethnic background. This analysis interestingly revealed that the homozygous wild type genotype (10/-10) was common in infertile men (77% - 47/61) and in normozoospermic control men (71.7% - 43/60). Our study failed to confirm any influence of the POLG gene polymorphism on the efficiency of the spermatogenesis.
Keywords: Infertility, normozoospermia, polymerase gamma
How to cite this article:
Poongothai J. No CAG repeat expansion of polymerase gamma is associated with male infertility in Tamil Nadu, South India. Indian J Hum Genet 2013;19:320-4 |
How to cite this URL:
Poongothai J. No CAG repeat expansion of polymerase gamma is associated with male infertility in Tamil Nadu, South India. Indian J Hum Genet [serial online] 2013 [cited 2016 May 24];19:320-4. Available from: http://www.ijhg.com/text.asp?2013/19/3/320/120823 |
 Introduction Introduction | |  |
Infertility affects every fifth couple in the fertile age. Male problems may contribute to the most common single defined cause of infertility. Recently, infertility has become a very big medical and social issue in India especially places where occupational hazards results in various health problems. The causes are known in less than half of these cases, out of which genetic or inherited disease and specific abnormalities in the Y chromosome are major factors. Accordingly the identification of genetic factors has become good practice for appropriate management of the infertile couple. [1] The three azoospermic regions of the Y chromosome represent genomic niches for spermatogenesis genes. [2] However, the search for causative mutations in the Y chromosome, autosomes, and X-linked genes has mostly been unsuccessful. Other polymorphisms are awaiting further confirmation, whereas for some (deleted in azoospermia like, ubiquitin specific protease 26, follicle stimulating hormone receptor) a lack of association with abnormal spermatogenesis has now been ascertained. [3]
In order to explore the causes of the unexplained infertility, there are several other candidate genes that are being studied that could lead to future breakthroughs. Quite recently scientific study of mitochondrial deoxyribonucleic acid (mtDNA) became possible and the role of mitochondrial dysfunction as cause of neurological disorders, optic neuropathy, infertility, and reproductive failure and many more abnormalities began to be explored. Sperm mitochondria play a significant role in spermatozoa functionality; therefore, genetic alterations of mtDNA may have consequences for normal fertilization. Mitochondria contain a single DNA polymerase, Polymerase gamma (POLG), [4] responsible for replication and repair of mtDNA. POLG gene encoding the catalytic subunit of POLG was mapped to the long arm of chromosome 15 (15q25) and includes a CAG repeat region. [4] A motif (CAG) 10 CAACAGCAG coding for a stretch of 13 glutamines is located in the first coding exon of this gene. From previous studies in humans, it has been established that the length of this CAG repeat is polymorphic with a major allele at 10 repeats. [5],[6] Ten copies of CAG repeat were found to be uniformly high (0.88) in different ethnic groups [7] and considered as the common allele, whereas the mutant alleles (not 10/- not 10 CAG repeats) were found to be associated with oligospermia/oligoasthenospermia in male infertility. [8] Various human pathologies have been ascribed to an expansion of a CAG repeat in some genes.
Recent data suggested the implication of POLG CAG repeats in infertility, [9] but are debated. [10] The search for causative mutations in the Y chromosome, and in autosomal and X-linked genes, has mostly been unsuccessful. As it is not clear whether the variable CAG genotype is the legitimate cause for male infertility, and as the likely significance of POLG CAG repeat polymorphisms has not been explored among Indian men, we have made an attempt to screen the CAG repeat motif of POLG gene exclusively in infertile men from two different regions of Tamil Nadu, South India.
 Materials and Methods Materials and Methods | |  |
Patient Selection
A total of 61 infertile men from two distinct regions namely, Erode and Nilgiri Districts of Tamil Nadu, South India, of which 45 were asthenozoospermic (> 60% of nonmotile sperms), and 16 were oligoasthenozoospermic (< 20 × 10 6 spermatozoa/ml; < 50% motile spermatozoa; and > 30% abnormal shape and size of spermatozoa) men and 60 normal males of proven fertility served as controls for this study. Further, these 61 infertile samples included 30 blood samples, 12 semen samples, and 19 paired samples; 60 normal controls included 45 blood, 11 semen, and four paired samples. All the procedures followed were in accordance with the ethical standards of Center for Cellular and Molecular Biology, Hyderabad.
A 10 ml of blood sample was collected from the individuals with ethylenediaminetetraacetic acid as anticoagulant. Semen samples were obtained by masturbation on two different occasions, separated by a 3-week interval, following a 3-day period of sexual abstinence. Semen samples were allowed to liquefy for 30 min at 37°C. Sperms in their ejaculates were analyzed for their motility, number, and morphology according to the guidelines of the World Health Organization, 1999. [11] Appropriate written informed consent was obtained from all participants in this study.
A team of urologists and andrologists had conducted a detailed clinical investigation and recorded the complete case histories. The samples were subjected to karyotyping and endocrinological assays for follicle-stimulating hormone, luteinizing hormone, testosterone, prolactin, and thyroid-stimulating hormone. Y chromosome microdeletion study was also performed for all the infertile men. [12] Only patients with an apparently normal 46, XY karyotype and who did not exhibit obstruction in the sperm delivery, endocrinological defect, pelvic injury, and Y chromosome abnormalities were included in this study.
Establishing the Identity of Paired Samples
Blood and semen samples (paired sample) of same patients (S37, SS37 and S41, SS41) were subjected to gene scan analysis to check for identity. On comparison of the short tandem repeats (STR) profile of the DNA samples [Figure 1] isolated from the blood and semen of same patients, it was pragmatic that both DNA profile were identical.
DNA Extraction and Polymerase Chain Reaction (PCR) for POLG CAG Repeats
DNA was extracted from 10 ml of peripheral blood [13] and from semen [14] using standard procedures. A pair of primers were used to amplify the region containing the CAG repeats, of which forward primer (5'-CCAGGGCCGGTTCCAGCTC-3') was designed, while the sequence of the reverse primer (mip31-5'GTGCTATCCTCGGAGGGCGGGCAGC) was obtained from elsewhere. [9] While synthesizing, forward primer was tagged with 5'-fluroscein amidite fluorescence dye in order to analyze the PCR products in the automated DNA analyzer (Applied Biosystems 3730).
DNA samples were analyzed for CAG repeat expansion by PCR in a 0.2-ml thin-wall tube under the following conditions: 5.0 ng of DNA, 1.2 mM MgCl 2 , 200 mm dNTPs, 10.0 pMol of each specific primer, and 1-2 U AmpliTaq Gold in a 10 μl reaction volume. Amplification was carried out in a MJ Research Thermal Cycler (Waltham, MA, USA) using the following cycling conditions: After an initial denaturation step at 94°C for 10 min, cycle parameters were 94°C for 71 min, 60°C for 45 s, and 72°C for 2 min for 30 cycles with a final extension of 72°C for 7 min. Amplified products were quantified by 2% agarose gel electrophoresis. The PCR products were then visualized under ultraviolet (UV ) light in transilluminator. On obtaining a single band devoid of any primer-dimer bands, the PCR products were then subjected to Genescan analysis [Figure 2]a.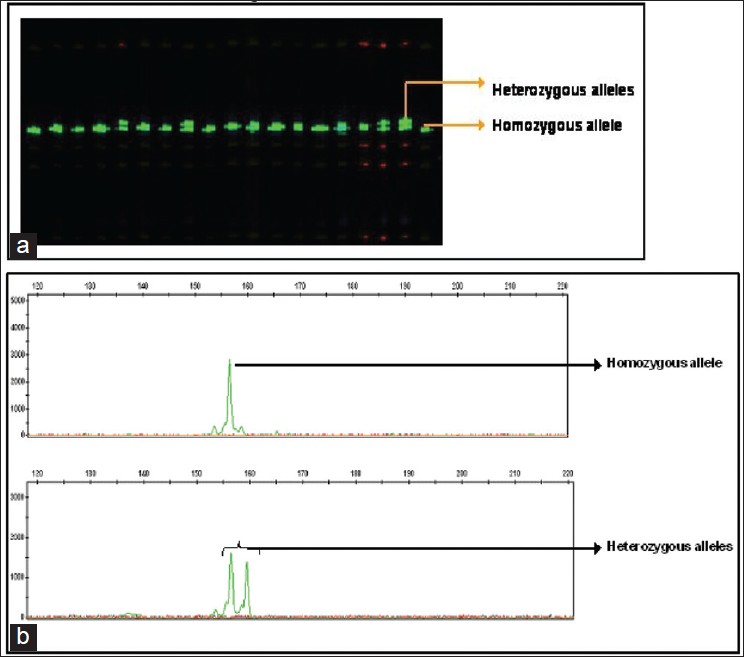 | Figure 2: (a) Gel picture showing CAG repeats of POLG gene (b) Gene scan electrophorograms showing CAG repeats of POLG gene
Click here to view |
Genotyping
A 1.0 μl of the PCR product was mixed with 0.3 μl of GS LIZ500 size standard and 8.7 μl of Hi-Di formamide and analyzed on 3730 DNA analyzer (ABI). The raw data were further analyzed using GeneMapper software (version 3.7) to ascertain the size of POLG CAG alleles [Figure 2]b. PCR and genotyping were repeated for all the samples to confirm the number of repeats.
Statistical analysis
We performed the χ2 test (Sys, version 10.0, from Statistical Package for Social Sciences Inc, Chicago, IL, USA) and odds ratio and obtained P-value (http://faculty.vassar.edu/lowry/VassarStats.html) to analyze the significance of the POLG polymorphism in male infertility.
 Results and Discussion Results and Discussion | |  |
Polymorphisms in CAG repeat number of the coding region (exon 1) of the catalytic subunit of the mtDNA POLG in 61 infertile men and 60 normozoospermic proven fertile control were analyzed to explore the possible associations between the POLG polymorphism and semen quality and fecundity. This analysis interestingly revealed that the common allele 10 (10 CAG repeats) was common in infertile and normozoospermic control men with a frequency of 77% in the infertile and 71.7% in the control group [Figure 3]. District-wise analysis of POLG CAG repeats in infertile men had shown almost the similar incidence [Figure 4]. The same tendency was observed when we looked at the allelic frequency of the common allele (10) in asthenozoospermic men (80%) and oligoasthenozoospermic men (68.8%) of Tamil Nadu State in South India. Allele 11 was the next common allele observed only in asthenozoospermic infertile men (2.2%) and 6.7% in normozoospermic men [Table 1].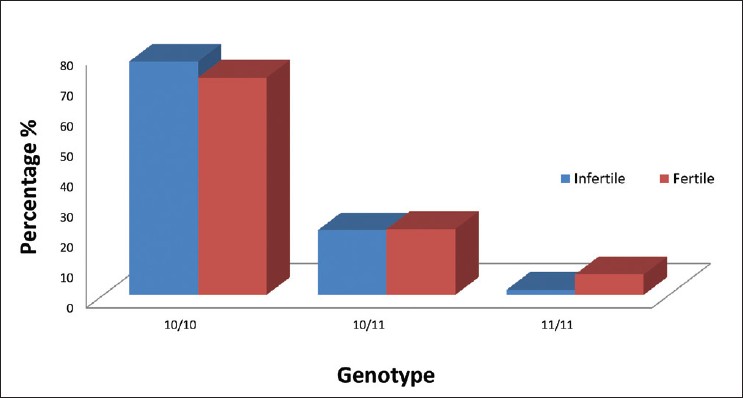 | Figure 3: Bar diagram indicating the genotype frequency distribution between infertile and control men of Erode and Nilgiri districts of Tamil Nadu
Click here to view |
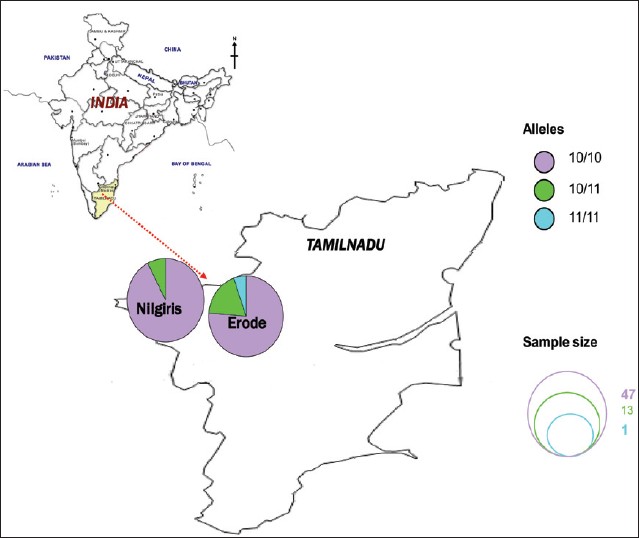 | Figure 4: Distribution of genotypic frequency of the CAG repeats of POLG gene in infertile men of Erode and Nilgiri districts of Tamil Nadu
Click here to view |
Narrow range of alleles and the genotypes were noticed in infertile and control men [Figure 3]; viz., two alleles (repeat length 10 to 11) and three genotypes [Figure 3] of which two (10/10, 11/11) were homozygous and the remaining one was heterozygous (10/11). Either blood or semen sample from each patient or both (paired sample) from same patient were preferred to confirm the results. No significance was found in statistical analysis of POLG genotypic frequency distribution between infertile men and normospermic men.
The search for new genetic hiccup represents one of the major tasks in present andrology, since majority of the idiopathic cases of male infertility are of genetic origin. The sole molecular genetic test that is routinely proposed in severe spermatogenic disturbances is screening for Yq microdeletion. Our preceding study on Y chromosome microdeletions revealed that it was responsible for 19.1% of male infertility. [12] Recently, the role of polymorphisms in genes potentially involved in spermatogenesis became of increasing interest. The most debated and controversial issue concerns the polymorphic CAG repeat-length variations of POLG gene. Mitochondrial POLG, is coded by POLG gene, which has a key role in spermatozoa functionality; therefore, genetic alterations of mtDNA may have consequences for normal fertilization. As the cause of spermatogenic failure was not clear in a large proportion of the infertile men studied, we intended to analyze the CAG repeat variations in the POLG gene of asthenozoospermic and oligoasthenospermic men. Interestingly, our study on 61 infertile men and 60 normal control men revealed occurrence of a very high frequency of the common allele (10/10) measuring 77% in the infertile and 71.7% in the control group. The same tendency was observed when we looked at the allelic frequency of the common allele (10) in asthenozoospermic men (80%) and oligoasthenozoospermic men (68.8%) of Tamil Nadu state in south India [Table 1]. The frequency of 10/11 heterozygous mutant allele [Table 1] and [Figure 3] was found to be slightly higher in infertile men than in the normozoospermic men. This observation supported the Amaral et al., 2007 [15] findings regarding existence of the oligoasthenoteratozoospermics (OAT) group with a significantly higher incidence of heterozygosity for CAG repeats. However, the biological significance of these heterozygous genotypes in male infertility remained unclear.
In concert with the previous report, [8],[10],[16] this study revealed that homozygotes with 10 repeats in both alleles (10/10) were the most common and constituted 77% of all infertile subjects which was almost equivalent to the normospermic control frequency (71.7%). Rovio et al., 1999; [7] have shown the frequent (88%) occurrence of genotype 10/10 in different ethnic group. As found by Krausz et al., 2004 and Aknin-Seifer et al., 2005; [17],[18] this study also suggested that the CAG repeat of the POLG gene does not play a significant role in men with severe male factor infertility. The previously reported variant of 10 repeats was found much more frequent in this study than any other allele. [5],[6] Another important observation made in this entire study was the absence of allele with more than 11 CAG repeats in infertile group. Only one 'homozygous mutants' out of 61 infertile patients was found, which was in disagreement with the data from Rovio et al., 2001; [8] where nine 'homozygous mutants' were identified out of 99 infertile men.
 Conclusion Conclusion | |  |
In conclusion, the present study confirmed no association between the POLG gene polymorphism and male infertility. Thus, if associated with infertility, the POLG gene polymorphism should be only considered as a minor possible contributing factor in infertile male patients with no impact on obtaining a pregnancy.
 References References | |  |
| 1. | Ferlin A, Arredi B, Foresta C. Genetic causes of male infertility. Rep Toxicol 2006;22:131-41. 
|
| 2. | Costa PN, Goncalves J, Plancha CE. The AZFc region of the Y chromosome: At the crossroads between genetic diversity and male infertility. Hum Reprod Update 2010;16:525-42. 
|
| 3. | Nuti F, Krausz C. Gene polymorphisms/mutations relevant to abnormal spermatogenesis. Reprod Biomed Online 2008;16:504-13. 
|
| 4. | Lim SE, Longley MJ, Copeland WC. The mitochondrial p55 accessory subunit of human DNA polymerase gamma enhances DNA binding, promotes processive DNA synthesis, and confers N-ethylmaleimide resistance. J Biol Chem 1999;274:38197-203. 
|
| 5. | Ropp PA, Copeland WC. Cloning and characterization of the human mitochondrial DNA polymerase, DNA polymerase gamma. Genomics 1996;36:449-58. 
|
| 6. | Lecrenier N, Foury F. New features of mitochondrial DNA replication system in yeast and man. Gene 2000;246:37-48. 
|
| 7. | Rovio A, Tiranti V, Bednarz AL, Suomalainen A, Spelbrink JN, Lecrenier N, et al. Analysis of the trinucleotide CAG repeat from the human mitochondrial DNA polymerase gene in healthy and diseased individuals. Eur J Hum Genet 1999;7:140-6. 
|
| 8. | Rovio AT, Marchington DR, Donat S, Schuppe HC, Abel J, Fritsche E, et al. Mutations at the mitochondrial DNA Polymerase (POLG) locus associated with male infertility. Nat Genet 2001;29:261-2. 
|
| 9. | Baklouti-Gargouri S, Ghorbel M, Chakroun N, Sellami A, Fakhfakh F, Ammar-Keskes L. The CAG repeat polymorphism of mitochondrial polymerase gamma (POLG) is associated with male infertility in Tunisia. Andrologia 2011. 
|
| 10. | Liu SY, Zhang CJ, Peng HY, Yao YF, Shi L, Chen JB, et al. CAG-repeat variant in the polymerase γ gene and male infertility in the Chinese population: A meta-analysis. Asian J Androl 2011;13:298-304. 
|
| 11. | World Health Organization. The World Health Report, WHO 1996; Geneva; 1996. p. 12. 
|
| 12. | Poongothai J, Manonayaki S. Y chromosome microdeletions in sperm DNA of infertile patients from Tamil Nadu, South India. IJU 2008;24:480-5. 
|
| 13. | Thangaraj K, Joshi MB, Reddy AG, Gupta NJ, Chakravarthy B, Singh L. CAG repeat expansion in androgen receptor gene is not associated with male infertility in Indian populations. J Androl 2002;23:815-8. 
|
| 14. | Thangaraj K, Joshi MB, Reddy AG, Rasalkar AA, Singh L. Sperm mitochondrial mutations as a cause of low sperm motility. J Androl 2003;24:388-92. 
|
| 15. | Amaral A, Ramalho-Santos J, St John JC. The expression of polymerase gamma and mitochondrial transcription factor A and the regulation of mitochondrial DNA content in mature human sperm. Hum Reprod 2007;22:1585-96. 
|
| 16. | Jensen M, Leffers H, Petersen JH, Nyboe Andersen A, Jorgensen N, Carlsen E, et al. Frequent polymorphism of the mitochondrial DNA polymerase gamma gene (POLG) in patients with normal spermiograms and unexplained subfertility. Hum Reprod 2004;19:65-70. 
|
| 17. | Krausz C, Guarducci E, Becherini L, Degl'Innocenti S, Gerace L, Balercia G, et al. The clinical significance of the POLG gene polymorphism in male infertility. J Clin Endocrinol Metab 2004;89:4292-7. 
|
| 18. | Aknin-Seifer IE, Touraine RL, Lejeune H, Jimenez C, Chouteau J, Siffroi JP, et al. Is the CAG repeat of mitochondrial DNA polymerase gamma (POLG) associated with male infertility? A multi-centre French study. Hum Reprod 2005;20:736-40. 
|
[Figure 1], [Figure 2], [Figure 3], [Figure 4]
[Table 1]
|






