|
 
 |
|
META ANALYSIS |
|
|
|
| Year : 2013 | Volume
: 19
| Issue : 4 | Page : 494-511 |
| |
Deoxyribonucleic acid repair gene X-ray repair cross-complementing group 1 polymorphisms and non-carcinogenic disease risk in different populations: A meta-analysis
Bagher Larijani1, Javad Mohammadi Asl2, Abbas Keshtkar1, Najmaldin Saki3, Fatemeh Ardeshir Larijani1, Fakher Rahim4
1 Department of Endocrinology and Metabolism, Endocrinology and Metabolism Research Center, Tehran University of Medical Sciences, Tehran, Iran
2 Department of Human and Medical Genetics, Toxicology Research Center, Ahvaz, Iran
3 Department of Hematology and Oncology, Thalassemia and Hemoglobinopathies Research Center, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran
4 Department of Molecular Medicine, Health Research Institute, Audiology Research Center, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran
| Date of Web Publication | 4-Jan-2014 |
Correspondence Address:
Fakher Rahim
PhD in molecular medicine, Toxicology Research Center, Ahvaz Jundishapur University of Medical Sciences, Ahvaz
Iran
 Source of Support: None, Conflict of Interest: None
PMID: 24497722 
 Abstract Abstract | | |
Purpose: This study aims to assess a meta-analysis of the association of X-ray repair cross-complementing group 1 (XRCC1) polymorphisms with the risk of various non-carcinogenic diseases in different population.
Materials and Methods: This meta-analysis was performed by critically reviewing reveals 38 studies involving 10043 cases and 11037 controls. Among all the eligible studies, 14 focused on Arg194Trp polymorphism, 33 described the Arg399Gln and three articles investigated on Arg280His. Populations were divided into three different ethnic subgroups include Caucasians, Asians and other (Turkish and Iranian).
Results: Pooled results showed no correlation between Arg194Trp and non-carcinogenic disease. There was only weak relation in the recessive (odds ratio [OR] =1.11, 95% confidence interval [CI]: 0.86-1.44) model in Asian population and dominant (OR = 1.04, 95% CI: 0.66-1.63) model of other populations. In Arg399Gln polymorphism, there was no relation with diseases of interest generally. In the pooled analysis, there were weak relation in the dominant (OR = 1.08, 95% CI: 0.86-1.35) model of Asian population and quite well-correlation with recessive (OR = 1.49, 95% CI: 1.19-1.88), dominant (OR = 1.23, 95% CI: 0.94-1.62), and additive (OR = 1.23, 95% CI: 0.94-1.62) models of other subgroup. For Arg280His, there was a weak relation only in the dominant model (OR = 1.06, 95% CI: 0.74-1.51).
Conclusion: The present meta-analysis correspondingly shows that Arg399Gln variant to be associated with increased non-carcinogenic diseases risk through dominant and recessive modes among Iranian and Turkish population. It also suggests a trend of dominant and recessive effect of Arg280His variant in all population and its possible protective effect on non-carcinogenic diseases.
Keywords: Arg194Trp, Arg280His, Arg399Gln, ethnicity, non-carcinogenic diseases, polymorphisms, X-ray repair cross-complementing group 1 gene
How to cite this article:
Larijani B, Asl JM, Keshtkar A, Saki N, Larijani FA, Rahim F. Deoxyribonucleic acid repair gene X-ray repair cross-complementing group 1 polymorphisms and non-carcinogenic disease risk in different populations: A meta-analysis. Indian J Hum Genet 2013;19:494-511 |
How to cite this URL:
Larijani B, Asl JM, Keshtkar A, Saki N, Larijani FA, Rahim F. Deoxyribonucleic acid repair gene X-ray repair cross-complementing group 1 polymorphisms and non-carcinogenic disease risk in different populations: A meta-analysis. Indian J Hum Genet [serial online] 2013 [cited 2016 May 24];19:494-511. Available from: http://www.ijhg.com/text.asp?2013/19/4/494/124385 |
 Introduction Introduction | |  |
There is increasing evidence suggests that damage to human deoxyribonucleic acid (DNA) might initiate the cancer, which caused by external agents such as chemical agents, ionizing radiation and ultraviolet (UV). [1],[2],[3] The X-ray repair cross-complementing group 1 (XRCC1) is a DNA repair gene and a number of its single nucleotide polymorphisms (SNPs) have been considered as a modifying risk factor for a variety of cancer types. Three different polymorphisms in XRCC1 gene have been identified at codon 399 (Arg to Gln), 194 (Arg to Trp) and 280 (Arg to His) until now, [4] which were predicted to be possibly damaging the XRCC1 function. [5]
The interactions of XRCC1 and its substrate result in assembly of the repair complex at the site of damage and regulate the activity of several repair enzymes. [6] The polymorphism Arg399Gln changes XRCC1's structure and maybe disrupt the combination of several repair enzymes, particularly poly (ADP-ribose) polymerase 1 (PARP1). Arg194Trp and Arg280His also change XRCC1's structure, but maybe not influence the function of XRCC1.
Previous analysis of case-control reports is the most predominant method of exploring the association between a specific gene and a disease. However, studies on XRCC1 polymorphisms in cancer have provided challenging and controversial results so far. Although other studies have found that the XRCC1 increase in breast cancer risk, [7],[8] and reports showed a possible protective effect, [9] while many studies observed no significant association between these polymorphisms and the disease. [10] Besides it was reported thatXRCC1 gene polymorphism is associated with several cancers including lung, esophageal, and prostate cancers, among different population. [11],[12],[13],[14],[15],[16]
Moreover, no evidence of any associations between Arg399Gln polymorphism and bladder cancer susceptibility has not shown, [17] hence other researchers reported that 399 Gln/Gln genotype is associated with a risk of lung cancer among Asians ethnicity, [18] and breast cancer in African Americans. [19] There are fairly few studies lead to observe the relationship between cancer risk and Arg280His variant up to the present time, only a single study revealed this association. [20],[21] Although, large numbers of epidemiologic studies have been evaluated the role of XRCC1 polymorphisms on various non-carcinogenic diseases, such as liver cirrhosis, Alzheimer, glaucoma, cataract, human immunodeficiency virus-1/acquired immunodeficiency syndrome, schizophrenia, type 2 diabetes [22],[23],[24],[25],[26],[27],[28],[29],[30],[31],[32],[33],[34],[35],[36],[37],[38],[39],[40],[41],[42],[43],[44],[45],[46],[47],[48],[49],[50],[51],[52],[53],[54],[55],[56] and cancers, but no such comprehensive analysis in the field of non-carcinogenic disease, is reported so far.
Nevertheless, a meta-analysis of all existing reports will help to create a more convincing result, because some of these studies were based on a small sample size, thus, subgroup analysis based on ethnic and other factors may also yield more meaningful results. It is important to perform a quantitative synthesis of the available evidence using more rigorous methods on the amounts of evidence have been accumulated so far. Therefore, we performed a meta-analysis of all eligible case-control studies published to date, to assess the association of XRCC1 polymorphisms with the risk of various non-carcinogenic diseases in different population.
 Materials and Methods Materials and Methods | |  |
Study selection
Relevant studies were identified in the PubMed, ISI web of science and Scopus using combinations of the search phrases "X-ray cross-complementing group 1," "polymorphism,'' "DNA repair gene" and all possible combination (the last search update on October 12, 2012). In addition, all publications in other databases such as IranMedex, scientific information database were searched. In a total of 383 retrieved relevant references, 38 publications were identified to be eligible for inclusion in the meta-analysis. These studies had a case-control study design that assessed the association between the XRCC1 Arg194Trp, the Arg399Gln and Arg280His polymorphisms and risk of non-carcinogenic diseases using human genomic DNA samples.
Inclusion criteria
Study design
Case-control studies were included in the evaluation, since this study design allows a comparison to be made between the affected individuals and healthy or disease-free ones, which is essential for the meta-analysis model.
Participants
Studies that included patients with any non-tumorigenic or non-carcinogenic condition were included in the evaluation.
Exclusion criteria
Studies that were not representative or not case-control were excluded. The studies that showed not enough data for analysis were excluded after contacting corresponding author twice.
Data extraction
Two reviewers independently screened all titles and abstracts. Full paper manuscripts of any titles/abstracts that appeared to be relevant were obtained where possible and the relevance of each study independently assessed by two reviewers according to the inclusion and exclusion criteria. Two authors (FR and NS) mined data and reached an agreement on all of the eligibility items, including author, journal and year of publication, location of study, selection and characteristics of cases and controls, control source, demographics, ethnicity and genotyping information.
Meta-analysis
The odds ratios (OR) of selected non-carcinogenic diseases associated with the XRCC1 Arg194Trp, the Arg399Gln and Arg280His polymorphisms were estimated for each study independently. We estimated the risk first for the variant homozygous genotypes, compared with the wild-type homozygous genotypes, assuming recessive and dominant effect models, respectively.
Statistical analysis
We calculated OR and 95% of confidence intervals (CI) to estimate non-carcinogenic risk associated with the XRCC1 polymorphism for each study. Inevitably, studies included in the meta-analysis differed in the variables of interest and thus, any kind of variability among studies may be termed heterogeneity. In meta-analysis, we examined the association between allele Trp of Arg194Trp and the risk of non-carcinogenic diseases compare with that of allele Arg, as well as using additive (Trp/Trp vs. Arg/Arg), recessive (Trp/Trp vs. [Arg/Trp + Arg/Arg]) and dominant ([Trp/Trp + Arg/Trp] vs. Arg/Arg) genetic models. The same method was applied to the other two polymorphisms. We evaluated the deviations from the Hardy-Weinberg equilibrium for the control group in each study by Chi-square test using a web-based program (http://www.ihg.gsf.de/cgi-bin/hw/hwa1.pl) for goodness of fit.
In the present study, both Der Simonian and Laird's random-effects method and Mantel-Haenszel's fixed-effects (FEs) method were used. In the meta-analysis, to evaluate the between-study heterogeneity both Chi-square-based Q-statistic and I-squared (I 2 ) tests were performed. Furthermore, according to Venice criteria, for the I 2 test included: <25% represents no heterogeneity, =25-50% represents moderate heterogeneity, =50-75% represents large heterogeneity and > 75% represents extreme heterogeneity. [57] So the heterogeneity was considered significant, if the P < 0.10 and I 2 > 25, a random-effect model was suitable, otherwise if the P ≥ 0.10and I 2 ≤ 25, a FE model was then used to estimate summary ORs and 95% CIs. Publication bias was assessed by a funnel plot based on the Egger's regression test and a t-test was implemented to determine the significance of the asymmetry. An asymmetric plot suggested possible publication bias (P ≥ 0.05 suggests no bias). All analyses were performed using STATA 11.0 (StataCorp LP, Lakeway Drive, College Station, Texas, USA). All the P values were two-sided.
 Results Results | |  |
Eligible studies
Thirty-nine reports focused on the role of any polymorphism of the XRCC1 gene in the non-carcinogenic risk were reviewed [Figure 1]. Four combined analysis include 3 individual case-control studies, two of which were also reported by Yousaf et al., [26] Ferguson et al., [45] and Olshan et al., [49] respectively. Thus, the present meta-analysis reveals 38 studies from 35 published papers involving 10043 cases and 11037 controls [Table 1]. Each sub-population study has treated as a separate in the analysis. Among all the eligible studies, 14 focused on Arg194Trp polymorphism, 33 described the Arg399Gln and 3 articles investigated on Arg280His. Populations were divided into three different ethnic subgroups include Caucasians, Asians, and other (Turkish and Iranian) [Table 1]. Considering each polymorphism, the overall genotype distributions in controls were significantly different (all P < 0.001) between Caucasian with Asian populations and other subgroup with Asian, but were not significant between Caucasian with other populations.
Arg194Trp
A total of 14 (3 Caucasian, 6 Asian, 5 other include Turkish) studies involving 3173 cases and 3863 controls addressed the association between Arg194Trp polymorphism and non-carcinogenic risk were reviewed [Table 2]. There was no between-study heterogeneity in ORs of individual studies of the recessive (χ2 = 9.21, I 2 = 0%, P = 0.757) and the additive (χ2 = 10.12, I 2 = 0%, P = 0.684) models, hence there was a moderate heterogeneity in the dominant model (χ2 = 19.80, I 2 = 34.4%, P = 0.100). Accordingly, we pooled the results using the FE model and found that TrpArg194Trp had a weak relation with non-carcinogenic disease in the recessive model [OR = 1.03, 95% CI: 0.88-1.22, [Figure 2]a, while used a FE model for the additive model [OR = 0.96, 95% CI: 0.79-1.17, [Figure 2]c and a random-effects model for the dominant type [OR = 0.94, 95% CI: 0.81-1.11, [Figure 2]c that showed no correlation likewise.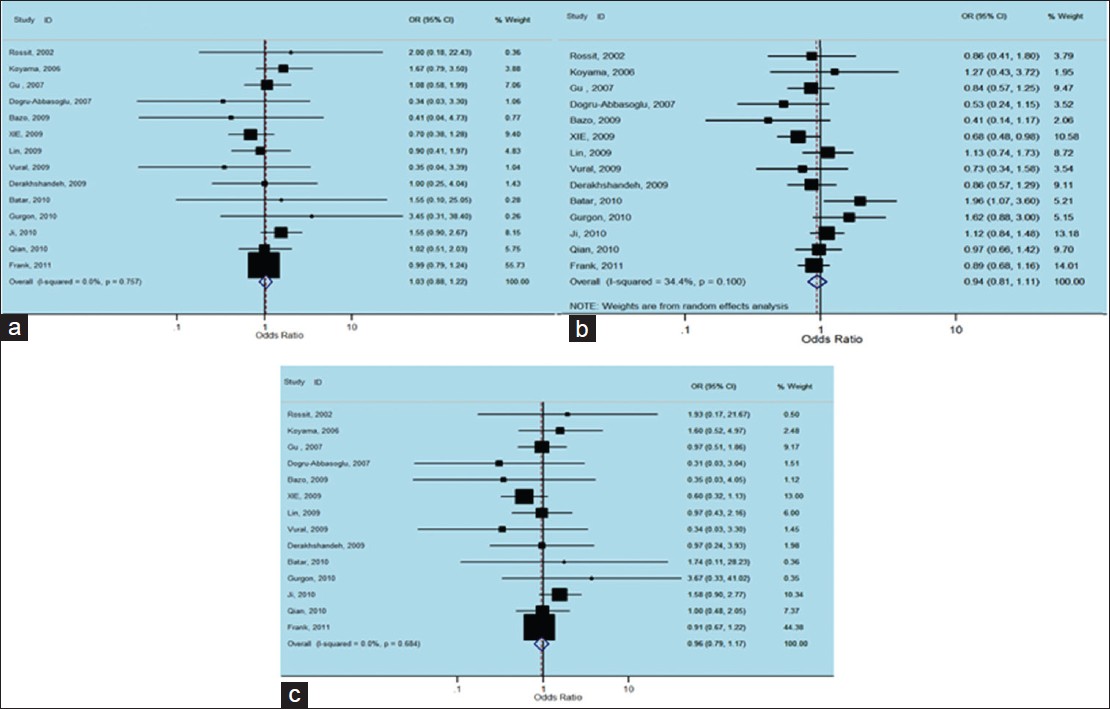 | Figure 2: Forest plots of odds ratios with 95% confidence interval for X - ray repair cross - complementing group 1 polymorphisms and risk of Non - carcinogenic disease. (a) Recessive model of Arg194Trp (Trp/Trp vs. Arg/Arg), (b) dominant model (Trp/Trp vs. Arg/Arg + Arg/Trp) and (c) additive model (Trp/Trp + Arg/Trp vs. Arg/Arg)
Click here to view |
Although we analyzing TrpArg194Trp polymorphism in stratified ethnic subgroups, there was no between-study heterogeneity in ORs of individual studies of the Caucasian subgroups in the recessive (χ2 = 0.82, I 2 = 0%, P = 0.664), the dominant (χ2 = 1.99, I 2 = 0%, P = 0.369) and the additive (χ2 = 0.95, I 2 = 0%, P = 0.621) models. Hence, we pooled the results using the FE analysis and found that TrpArg194Trp was not related with non-carcinogenic disease in the recessive (OR = 0.99, 95% CI: 0.79-1.23, [Figure 3]a, dominant (OR = 0.85, 95% CI: 0.67-1.08, [Figure 3]b and additive (OR = 0.90, 95% CI: 0.67-1.21, [Figure 3]c models. Meanwhile, when we analyzed the Asian subgroups, there was no between-study heterogeneity in ORs of individual studies of the recessive (χ2 = 5.11, I 2 = 2.2%, P = 0.402), the dominant (χ2 = 5.75, I 2 = 13.1%, P = 0.331) and the additive (χ2 = 5.64, I 2 = 11.3%, P = 0.343) models. Thus, we pooled the results using the FE analysis and found that TrpArg194Trp was not related with non-carcinogenic disease in dominant (OR = 0.95, 95% CI: 0.81-1.11, [Figure 3]e, but had a weak correlation with the recessive (OR = 1.11, 95% CI: 0.86-1.44, [Figure 3]d and the additive (OR = 1.04, 95% CI: 0.79-1.37, [Figure 3]f models. Then in the analysis of the other subgroups, there was no between-study heterogeneity in ORs of individual studies of the recessive (χ2 = 2.75, I 2 = 0%, P = 0.600), and the additive (χ2 = 3.09, I 2 = 0%, P = 0.543) models, but there was a large heterogeneity in the dominant (χ2 = 10.78, I 2 = 62.9%, P = 0.029), so we took a random-effects analysis. Consequently we pooled the results using the FE analysis and found that TrpArg194Trp was not related with non-carcinogenic disease in the recessive [OR = 0.86, 95% CI: 0.36-2.03, [Figure 3]g and additive [OR = 0.85, 95% CI: 0.38-2.00, [Figure 3]i models, while had a weak relation with the dominant [OR = 1.04, 95% CI: 0.66-1.63, [Figure 3]h using random-effect analysis.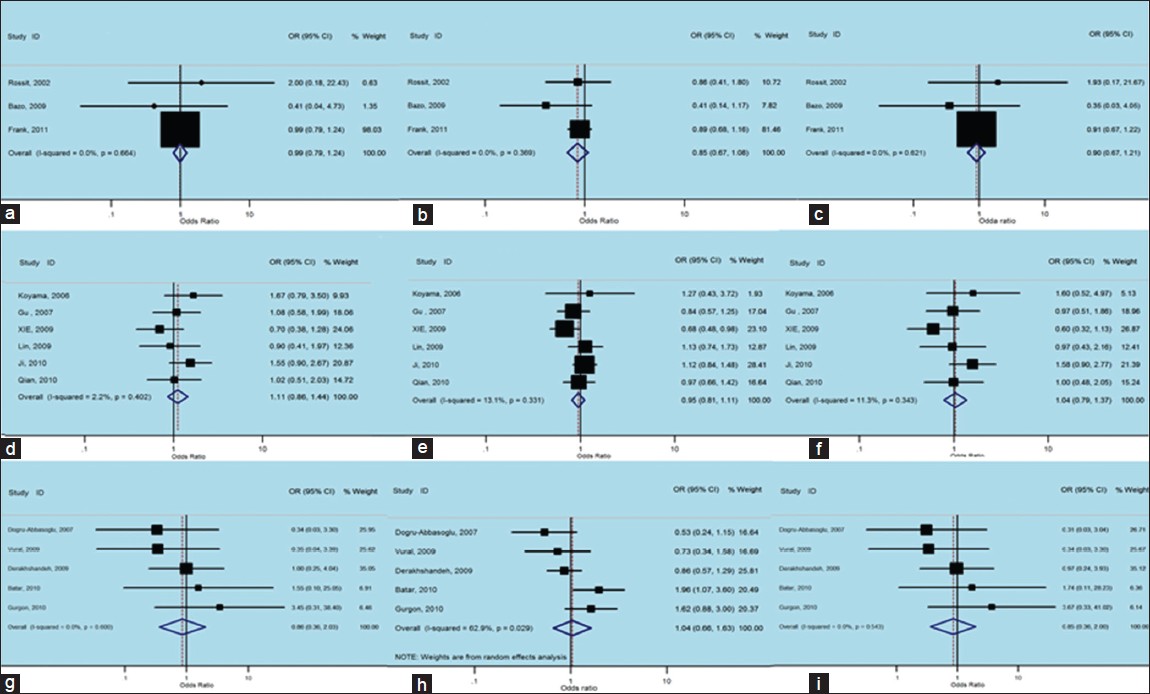 | Figure 3: Forest plots of odds ratios with 95% confidence interval for X - ray repair cross - complementing group 1 polymorphisms and risk of non - carcinogenic disease (right) recessive model of Arg194Trp (Trp/Trp vs. Arg/Arg), (middle) dominant model (Trp/Trp vs. Arg/Arg+ Arg/Trp) and (left) additive model (Trp/Trp + Arg/Trp vs. Arg/Arg); first row is a subgroup analysis in Caucasian population under an fixed - effects (FEs) model (a - c); second row is a subgroup analysis in Asian population under an FEs model (d - f); third row is a subgroup analysis as other population under an FEs model (g and i) and random-effects
Click here to view |
Arg399Gln
There were 33 studies (3099 cases and 3169 controls) concerning eight Caucasian, 14 Asian and 11 other subgroups, which addressed the relation of XRCC1 Arg399Gln polymorphism and the risk of non-carcinogenic diseases. We examined the association between Arg399Gln XRCC1 polymorphism and non-carcinogenic diseases risk, assuming various inheritance models of the399Gln allele for each individual study [Table 3]. There was a large between-study heterogeneity in ORs of individual studies of the recessive (χ2 = 72.27, I 2 = 55.7%, P = 0.000) and the additive (χ2 = 56.18, I 2 = 43.0%, P = 0.005) models, but a moderate heterogeneity in the dominant model (χ2 = 74.18, I 2 = 56.9%, P = 0.000). Hence, we pooled the results using the random-effect analysis and found that Gln Arg399Gln has a weak relation with non-carcinogenic disease in the recessive [OR = 1.02, 95% CI: 0.86-1.21, [Figure 4]a, additive [OR = 1.15, 95% CI: 0.96-1.39, [Figure 4]c and the dominant [OR = 1.10, 95% CI: 0.96-1.26, [Figure 4]b models.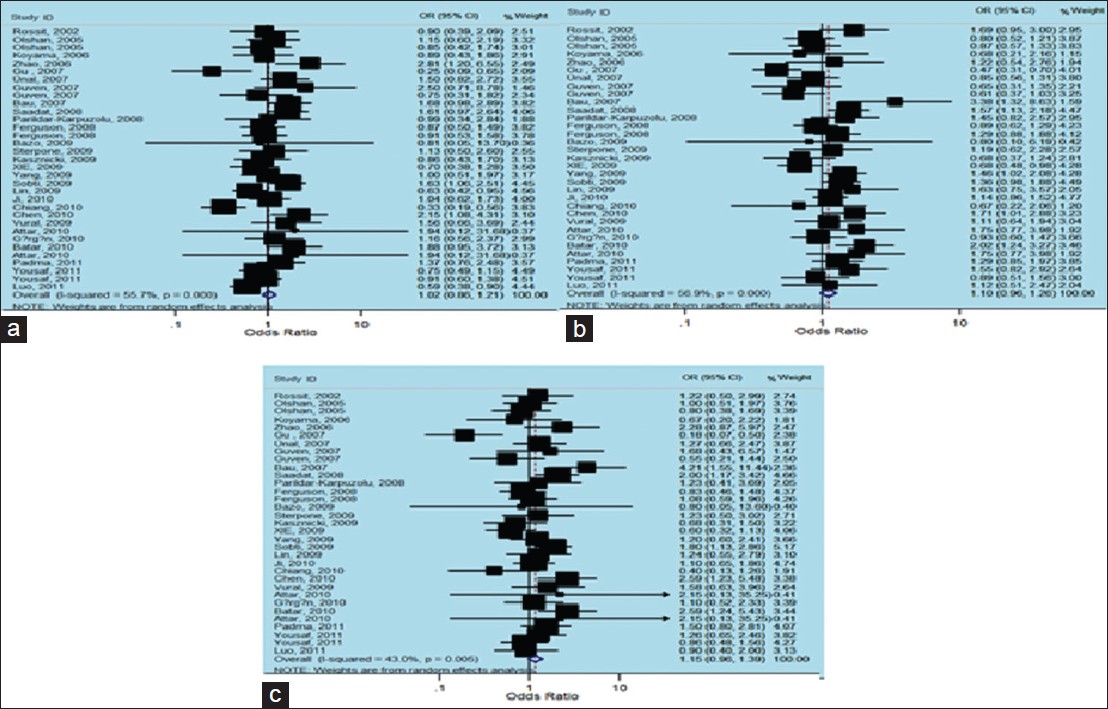 | Figure 4: Forest plots of odds ratios with 95% confidence interval for X - ray repair cross - complementing group 1 polymorphisms and risk of non - carcinogenic disease. (a) Recessive model of Arg399Gln (Gln/Gln vs. Arg/Arg), (b) dominant model (Gln/Gln vs. Arg/Arg + Arg/Gln) and (c) additive model (Gln/Gln + Arg/Gln vs. Arg/Arg)
Click here to view |
There was no between-study heterogeneity in ORs of individual studies of the Caucasian subgroups in the recessive (χ2 = 0.83, I 2 = 0%, P = 0.997), the dominant (χ2 = 8.73, I 2 = 19.8%, P = 0.273) and the additive (χ2 = 1.92, I 2 = 0%, P = 0.964) models. So we pooled the results using the FE analysis and found that Gln Arg399Gln was not related with non-carcinogenic disease in the recessive [OR = 0.93, 95% CI: 0.73-1.20, [Figure 5]a, dominant [OR = 0.99, 95% CI: 0.84-1.18, [Figure 5]b and additive [OR = 0.94, 95% CI: 0.72-1.22, [Figure 5]c models. Furthermore, when we analyzed the Asian subgroups, there was a large between-study heterogeneity in ORs of individual studies of the recessive (χ2 = 50.82, I 2 = 74.4%, P = 0.000), the dominant (χ2 = 35.89, I 2 = 63.8%, P = 0.001) and the additive (χ2 = 33.36, I 2 = 61.0%, P = 0.002) models. Hence, we pooled the results using the random-effect analysis and found that Gln Arg399Glnwas not related with non-carcinogenic disease in the recessive [OR = 0.88, 95% CI: 0.66-1.18, [Figure 5]d, while it presented a weak correlation with dominant [OR = 1.08, 95% CI: 0.86-1.35, [Figure 5]e, and additive [OR = 1.05, 95% CI: 0.77-1.43, [Figure 5]f models. Then in the analysis of the other subgroups, there was no between-study heterogeneity in ORs of individual studies of the recessive (χ2 = 0.40, I 2 = 0%, P = 0.819), and the dominant (χ2 = 0.22, I 2 = 0%, P = 0.898) models, but there was a large heterogeneity in the additive (χ2 = 5.03, I 2 = 60.2%, P = 0.081), so we took a random-effects analysis. Therefore, we pooled the results using the FE model and found that TrpArg194Trp was related with non-carcinogenic disease in the recessive [OR = 1.49, 95% CI: 1.19-1.88, [Figure 5]g, and additive [OR = 1.61, 95% CI: 1.24-2.10, [Figure 5]i models, using random-effects analysis, it was correlated with the dominant [OR = 1.23, 95% CI: 0.94-1.62, [Figure 5]h model as well. | Figure 5: Forest plots of odds ratios with 95% confidence interval for X - ray repair cross - complementing group 1 polymorphisms and risk of non - carcinogenic disease (right) recessive model of Arg399Gln (Gln/Gln vs. Arg/Arg), (middle) dominant model (Gln/Gln vs. Arg/Arg+ Arg/Gln) and (left) Additive model (Gln/Gln + Arg/Gln vs. Arg/Arg); first row is a subgroup analysis in Caucasian population under an fixed - effects (FEs) model (a - c); second row is a subgroup analysis in Asian population under an FEs model (d - f); third row is a subgroup analysis in other population under an FEs model (g-i)
Click here to view |
Arg280His
There were only three studies (1115 cases and 815 controls) involving one Caucasian and 2 Asian reports, that investigating the relation of XRCC1 Arg280His polymorphism and the risk of non-carcinogenic disease. We examined the relationship between Arg280His XRCC1 polymorphism and non-carcinogenic diseases risk, assuming various inheritance models of the 280His allele for each individual study [Table 4]. There was no between-study heterogeneity in ORs of individual studies of the recessive (χ2 = 0.40, I 2 = 0%, P = 0.819) and the additive (χ2 = 0.22, I 2 = 0%, P = 0.898) models, whereas the dominant model has a large heterogeneity (χ2 = 5.03, I 2 = 60.2%, P = 0.081). Accordingly we pooled the results using the FE analysis in the recessive [OR = 0.50, 95% CI: 0.22-1.11, [Figure 6]a, additive [OR = 0.58, 95% CI: 0.19-1.74, [Figure 6]c and using random-effects analysis in the dominant models [OR = 1.06, 95% CI: 0.74-1.51, [Figure 6]b and found that His Arg280His was not related with non-carcinogenic disease.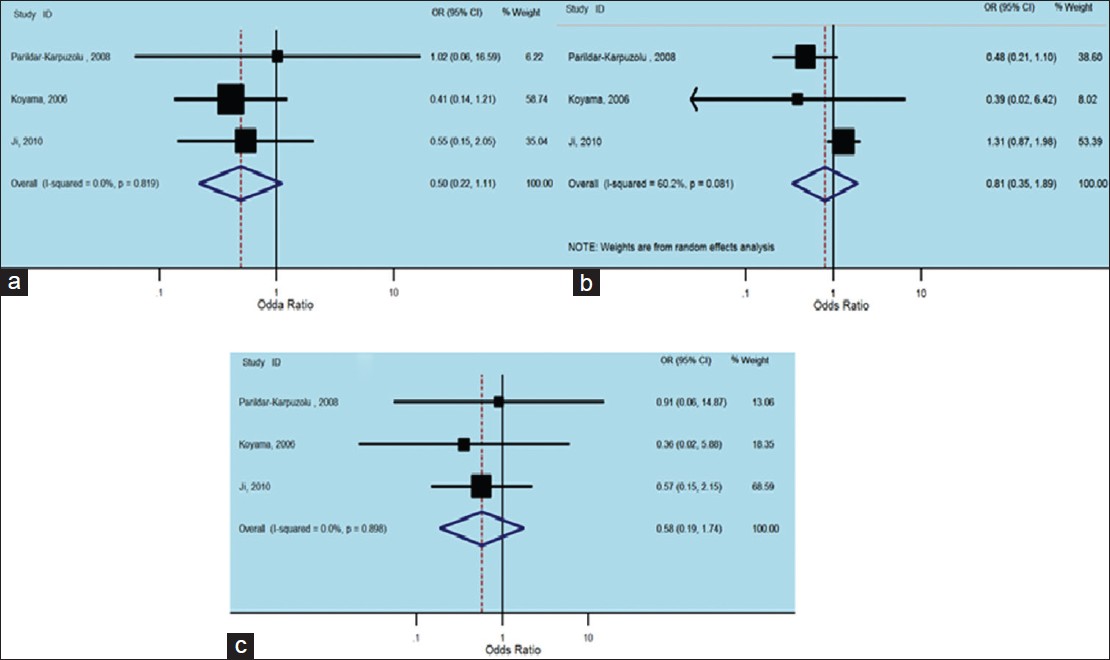 | Figure 6: Forest plots of odds ratios with 95% confidence interval for X - ray repair cross - complementing group 1 polymorphisms and risk of Non - carcinogenic disease. (a) Recessive model of Arg280His (His/His versus Arg/Arg), (b) dominant model (His/His vs. Arg/Arg + Arg/His) and (c) additive model (His/His + Arg/His vs. Arg/Arg)
Click here to view |
Sensitivity analysis
We implemented sensitivity analyses to assess the effect of those studies that are not in Horner-Wadsworth-Emmons. [28],[36],[38],[44] The results stayed similar when eliminating those studies. The present analyses of hospital based and population-based studies individually also did not lead to a different conclusion. In addition, meta-regression did not find a significant difference between various study designs.
Publication bias
Funnel plots and Egger's test were performed to assess publication bias, which suggested that there were no publication bias for the comparison of Arg399Gln polymorphism, in term of recessive (t = 1.07, P = 0.294), dominant (t = 0.39, P = 0.701) and additive (t = −0.57, P = 0.575) models [Figure 7] and [Table 5]. Furthermore, there were no publication bias for the comparison of Arg194Trp polymorphism, in term of recessive (t = −0.01, P = 0.995), dominant (t = −0.19, P = 0.854) and additive (t = −0.12, P = 0.910) models [Figure 9] and [Table 5]. Besides, there were no publication bias for the comparison of Arg280His polymorphism, in term of recessive (t = 3.13, P = 0.197), dominant (t = −1.08, P = 0.475) and additive (t = −0.00, P = 0.997) models [Figure 11] and [Table 5]. However, when we stratified Arg399Gln, Arg194Trp and Arg280His polymorphisms, according to different ethnic subgroups include Caucasian, Asian and other; there was no public bias in each subgroup [Figure 8], [Figure 10], [Figure 12] and [Table 5] and [Table 6].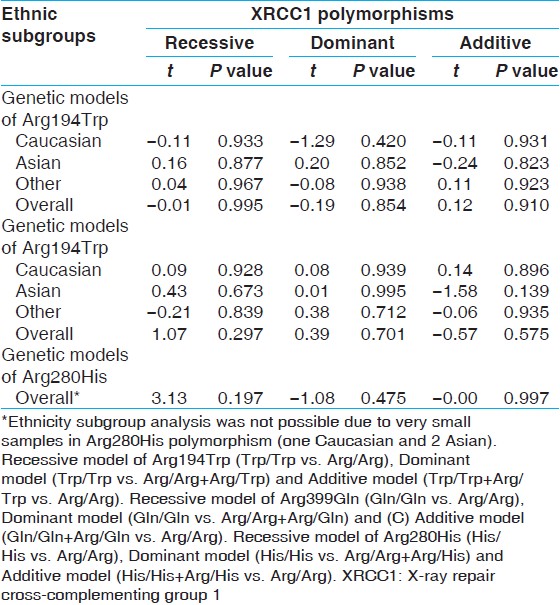 | Table 5: Egger's test variables to assess publication bias and comparison of 399Gln versus 399Arg, 194Trp versus 194Arg and 280His versus 280Arg
Click here to view |
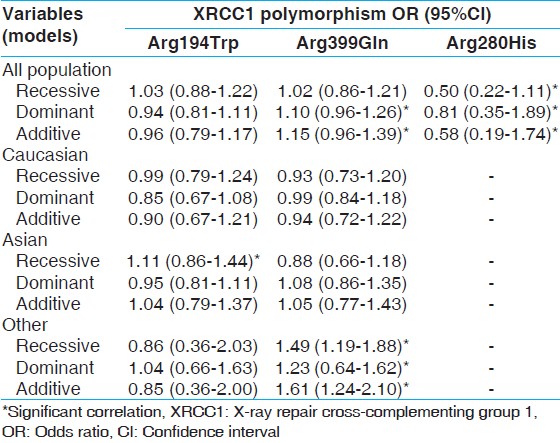 | Table 6: The association of XRCC1 gene polymorphisms and non-carcinogenic risk by assuming different population
Click here to view |
 | Figure 7: Begg's funnel plot of the Egger's test of allele comparison for publication bias. (a) Additive model of Arg194Trp (Trp/Trp vs. Arg/Arg), (b) dominant model (Trp/Trp vs. Arg/Arg+ Arg/Trp) and (c) recessive model (Trp/Trp + Arg/Trp vs. Arg/Arg)
Click here to view |
 | Figure 8: Begg's funnel plot of the Egger's test of allele comparison for publication bias (top) additive model of Arg194Trp (Trp/Trp vs. Arg/Arg), (middle) dominant model (Trp/Trp vs. Arg/Arg+ Arg/Trp) and (bottom) recessive model (Trp/Trp + Arg/Trp vs. Arg/Arg); first row is a subgroup analysis in Caucasian population (a - c); second row is a subgroup analysis in Asian population (d - f); Third row is a subgroup analysis in other population (g - i)
Click here to view |
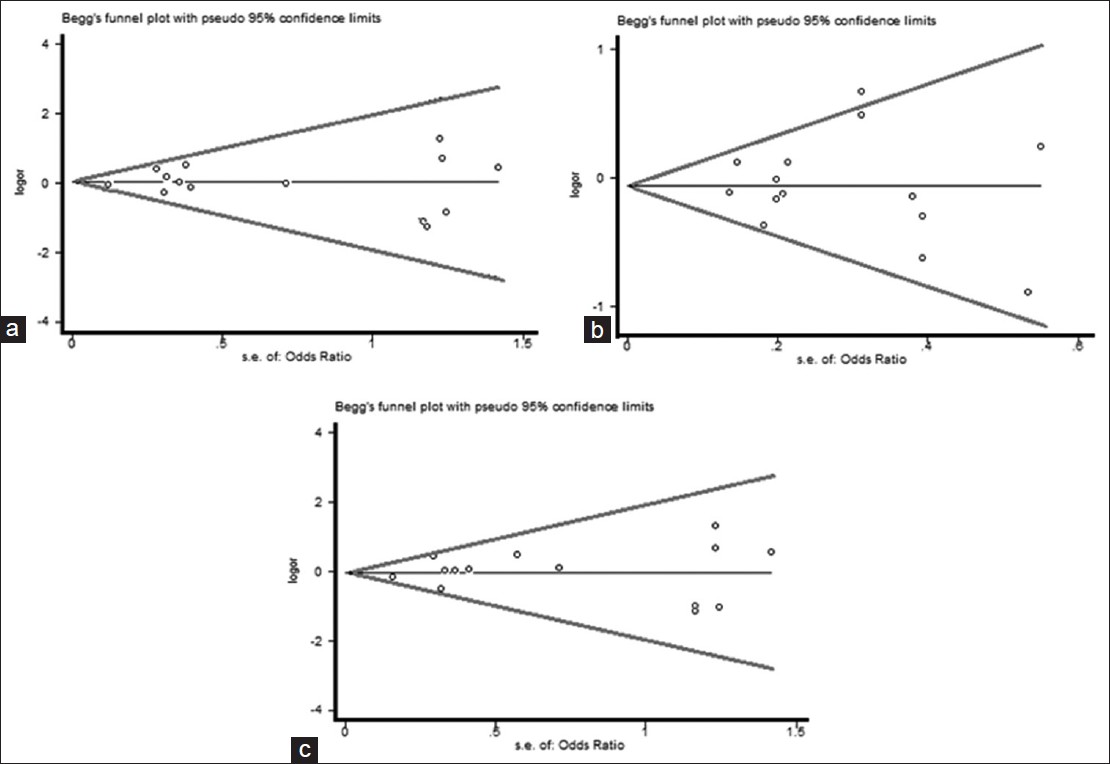 | Figure 9: Begg's funnel plot of the Egger's test of allele comparison for publication bias (top) (right) additive model of Arg399Gln (Gln/Gln vs. Arg/Arg), (middle) dominant model (Gln/Gln vs. Arg/Arg+ Arg/Gln) and (bottom) recessive model (Gln/Gln + Arg/Gln vs. Arg/Arg)
Click here to view |
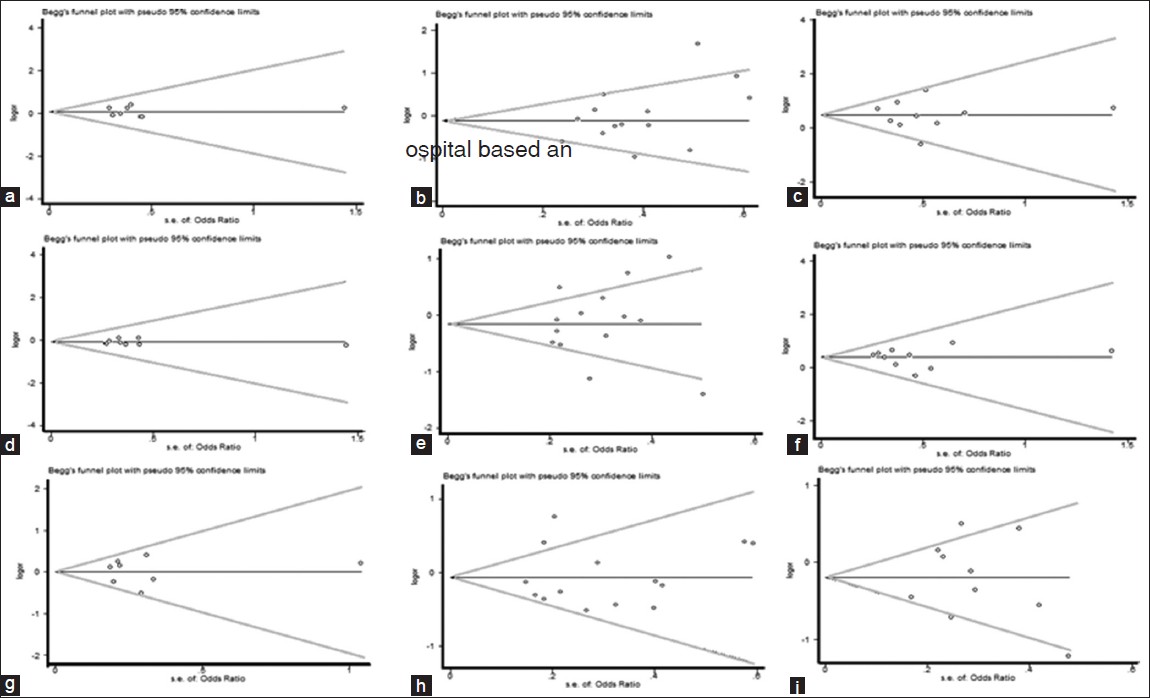 | Figure 10: Begg's funnel plot of the Egger's test of allele comparison for publication bias (top) (right) additive model of Arg399Gln (Gln/Gln vs. Arg/Arg), (middle) dominant model (Gln/Gln vs. Arg/Arg + Arg/Gln) and (bottom) Recessive model (Gln/Gln + Arg/Gln versus Arg/Arg); First row is a subgroup analysis in Caucasian population (a - c); second row is a subgroup analysis in Asian population (d - f); third row is a subgroup analysis in other population (g - i)
Click here to view |
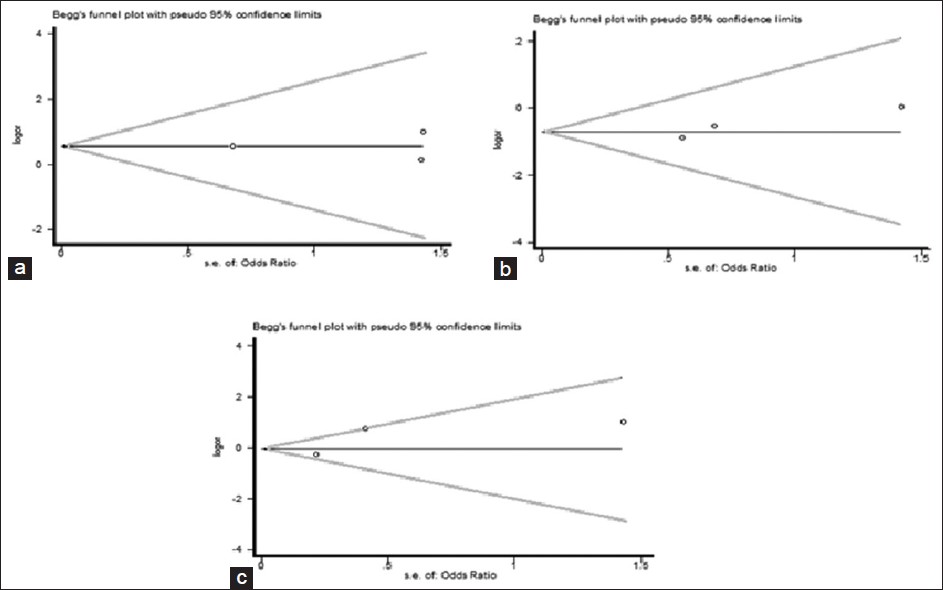 | Figure 11: Begg's funnel plot of the Egger's test of allele comparison for publication bias. (a) Additive model of Arg280His (Gln/Gln vs. Arg/Arg), (His/His vs. Arg/Arg), (b) dominant model (His/His vs. Arg/Arg + Arg/His) and (c) additive model (His/His + Arg/His vs. Arg/Arg)
Click here to view |
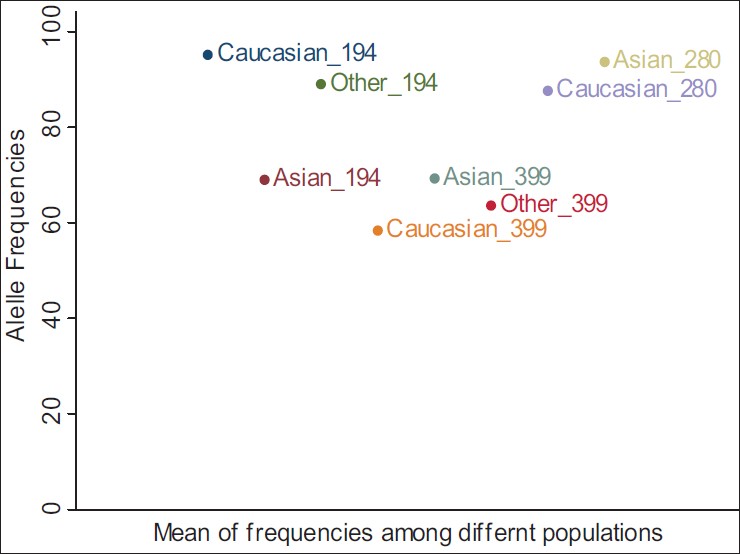 | Figure 12: Mean of Arg allele frequencies for non-carcinogenic diseases between different populations
Click here to view |
 Discussion Discussion | |  |
Large and unbiased molecular and genetic epidemiologic studies of SNPs such as DNA repair genes, may provide insight into the in vivo relations between the candidate genes and non-carcinogenic and cancer risk. XRCC1 is very important repair gene for efficient base excision and single-strand break in DNA. The present meta-analysis observed Arg194Trp, Arg280His and Arg399Gln polymorphisms of the XRCC1 gene and their associations with non-carcinogenic disease risk in various populations and ethnicity, by critically reviewing 38 studies.
Many of the studies indicated the association between the oxidative or UV light DNA damage and cataract development, [58],[59],[60],[61],[62] that the contribution of DNA damage in cataract pathogenesis indicate the role of DNA repair enzymes such as XRCC1. An epidemiologic study that reviewed twenty-two researches revealed a well-documented risk for cataract and DNA damage due to UV exposure. [63] Previous studies showed no association between Arg194Trp polymorphism and indicators of DNA repair capacity, such as, sensitivity to ionizing radiation or DNA-adduct levels. [64] Hence, our meta-analysis found evidence that 194Trp variant altered non-carcinogenic disease risk among Asian populations. However, other studies showed that this polymorphism exhibited significantly lower values of chromosomal breaks per cell and the protective effect of 194Trp. [65],[66] Studies suggest that Arg194Trp polymorphism does not modify the risk for non-carcinogenic disease including alcoholic cirrhosis, pre-eclampsia (PE) and idiopathic azoospermia in Asian, Caucasian and other population, [24],[32],[42] while some studies showed a protective effect against other disease such as chronic obstructive pulmonary disease (COPD) and Pterygiumin Asian population. [43],[53] In some meta-analysis about the association between Arg194Trp and risk of cancer considering different genetic models, no evidence of the protective effect against the bladder and breast cancer has been found in Asian and Caucasian. [17],[67],[68],[69] However, others showed Arg280His genotype increased risk for differentiated thyroid carcinoma and gastric cardiac adenocarcinoma in the dominant model, while mildly reduced the risk for this cancer in Asian and Other (Iranian) population. [70],[71] Our meta-analysis also recommends a tendency towards recessive mode of risky effect of 194Trp, which suggest that further studies should be performed to evaluate the effect of this polymorphism.
Moreover, for XRCC1-Arg399Gln polymorphism studies showed that this polymorphism may modify the risk for the non-carcinogenic disease including alcoholic cirrhosis, PE, Alzheimer's disease (AD), ocular diseases include primary open angle glaucoma, cataract, Pterygium, severe chronic atrophic gastritis and idiopathic azoospermia in Asian, Caucasian and other population, [23],[24],[27],[29],[30],[32],[42],[43],[68] while some studies showed no association with other disease such as COPD and endometriosis in Asian and other population. [31],[53] Several well-known atherosclerotic risk factors, such as dyslipidemia and diabetes mellitus, lead to DNA damage, [69] thus the effects of this risk factors on DNA damage in coronary artery disease (CAD) have been demonstrated formerly [70],[71] and found no associations between CAD and Arg399Gln polymorphism in other (Turkey) population [34] whereas, other study showed a relationship between CAD and Arg399Gln, polymorphisms in Caucasian. [35] In cystic fibrosis, there was slight correlation between Arg399Gln polymorphism with liver status and pancreatic insufficiency in Caucasian, but this correlation was not significant. [36] In a meta-analysis of Asian (Taiwanese Han Chinese) and Caucasian (Brazilian, and Polish) populations showed that the XRCC1 (Arg399Gln polymorphism) was associated with systemic lupus erythematous incidence. [40] Furthermore, the XRCC1 (Arg399Gln polymorphism) may affect risk of two major birth defects including spina bifida and oral clefts in Caucasian (USA) population. [49] The majority of studies have reported that there was no association between the XRCC1 (codon 399) polymorphism and cancer. [72],[73],[74],[75],[76],[77],[78],[79] In the minority of researches, a weak but statistically significant association has been found in Asian countries, entirely. [18],[72],[73],[74] Our meta-analysis suggests that 399Gln increases non-carcinogenic disease risk by 50%, 25% and 60% with recessive, dominant and additive models in other population only, respectively, which indicated that the genotype distributions of Arg399Gln varied with ethnicity. There may be two explanations concerning the difference in results. Genetic, environmental, and ethnic differences in allele frequency for the investigated polymorphisms can affect results in studies. One possible explanation could be differences in ethnicity in term of dietary habits and drinking, health-care access and socioeconomic factors. Another more reasonable clarification may be linked to diversity in linkage or genetic associations between alleles in different populations, which formerly were reported in cancer. [80]
From the Biological point of view, 280His codon is placed in the proliferating cell nuclear antigen-binding region. Previously, it was suggested 280His codon to be associated with higher bleomycin sensitivity, which resulted in a reduced DNA repair capacity produced by bleomycin. [71] Studies showed that XRCC1-Arg280His polymorphism had a protective effect on non-carcinogenic disease such as AD, rheumatoid arthritis in other (Turkish) and Asian (Taiwanese and Japanese) population, [23],[46],[61] while does not meet the frequency criteria for being considered an important SNP in some non-carcinogenic disease like ocular disease (Pterygium), severe chronic atrophic gastritis, spina bifida and oral clefts among Asian (Chinese) and Caucasian (Irish and American) population. [38],[44],[45],[49] Our meta-analysis suggests a tendency for Asian and Caucasian populations harboring Arg280His to have a protective effect against non-carcinogenic disease through both recessive and dominant effect [Table 5]. These varying effects in Asian and Caucasian populations may be due to the difference in distributions of this SNP, with a lower frequency in Caucasian population (4-6%) when compared with Asian population [Table 4]. As studies of Arg280His among all populations especially Asian and other subgroup are at present in adequate, further studies including a broader variety of Asian and other subgroup subjects should be carried out to approve whether this XRCC1 variant alters non-carcinogenic disease risk differently in Asian and other subgroup populations.
 Conclusion Conclusion | |  |
The present meta-analysis correspondingly shows that comprising diverse population is very important since susceptibility loci might vary indifferent ethnic groups. To ratify our findings, widespread studies with enlarged sample size and various populations are essential to explain the role of all polymorphism ofXRCC1 genes in the pathogenesis of non-carcinogenic diseases. Finally, our meta-analysis showed Arg399Gln variant to be associated with increased non-carcinogenic diseases risk through dominant and recessive modes among Iranian and Turkish population. It also suggests a trend of dominant and recessive effect of Arg280His variant in all population and its possible protective effect on non-carcinogenic diseases as well.
 References References | |  |
| 1. | Rastogi RP, Richa, Kumar A, Tyagi MB, Sinha RP. Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair. J Nucleic Acids 2010;2010:592980. 
|
| 2. | Okayasu R. Repair of DNA damage induced by accelerated heavy ions - A mini review. Int J Cancer 2012;130:991-1000. 
|
| 3. | Carpenter DO, Arcaro KF, Bush B, Niemi WD, Pang S, Vakharia DD. Human health and chemical mixtures: An overview. Environ Health Perspect 1998;106 Suppl 6:1263-70. 
|
| 4. | Shen MR, Jones IM, Mohrenweiser H. Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res 1998;58:604-8. 
|
| 5. | Metsola K, Kataja V, Sillanpää P, Siivola P, Heikinheimo L, Eskelinen M, et al. XRCC1 and XPD genetic polymorphisms, smoking and breast cancer risk in a Finnish case-control study. Breast Cancer Res 2005;7:R987-97. 
|
| 6. | Caldecott KW, Aoufouchi S, Johnson P, Shall S. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular 'nick-sensor' in vitro. Nucleic Acids Res 1996;24:4387-94. 
|
| 7. | Saadat M, Kohan L, Omidvari S. Genetic polymorphisms of XRCC1 (codon 399) and susceptibility to breast cancer in Iranian women, a case-control study. Breast Cancer Res Treat 2008;111:549-53. 
|
| 8. | Loizidou MA, Michael T, Neuhausen SL, Newbold RF, Marcou Y, Kakouri E, et al. Genetic polymorphisms in the DNA repair genes XRCC1, XRCC2 and XRCC3 and risk of breast cancer in Cyprus. Breast Cancer Res Treat 2008;112:575-9. 
|
| 9. | Costa S, Pinto D, Pereira D, Rodrigues H, Cameselle-Teijeiro J, Medeiros R, et al. DNA repair polymorphisms might contribute differentially on familial and sporadic breast cancer susceptibility: A study on a Portuguese population. Breast Cancer Res Treat 2007;103:209-17. 
|
| 10. | Thyagarajan B, Anderson KE, Folsom AR, Jacobs DR Jr, Lynch CF, Bargaje A, et al. No association between XRCC1 and XRCC3 gene polymorphisms and breast cancer risk: Iowa Women's Health Study. Cancer Detect Prev 2006;30:313-21. 
|
| 11. | Lee JM, Lee YC, Yang SY, Yang PW, Luh SP, Lee CJ, et al. Genetic polymorphisms of XRCC1 and risk of the esophageal cancer. Int J Cancer. 2001 Jul 20; 95:240-6. 
|
| 12. | Lee SG, Kim B, Choi J, Kim C, Lee I, Song K. Genetic polymorphisms of XRCC1 and risk of gastric cancer. Cancer Lett 2002;187:53-60. 
|
| 13. | van Gils CH, Bostick RM, Stern MC, Taylor JA. Differences in base excision repair capacity may modulate the effect of dietary antioxidant intake on prostate cancer risk: An example of polymorphisms in the XRCC1 gene. Cancer Epidemiol Biomarkers Prev 2002;11:1279-84. 
|
| 14. | Misra RR, Ratnasinghe D, Tangrea JA, Virtamo J, Andersen MR, Barrett M, et al. Polymorphisms in the DNA repair genes XPD, XRCC1, XRCC3, and APE/ref-1, and the risk of lung cancer among male smokers in Finland. Cancer Lett 2003;191:171-8. 
|
| 15. | Moullan N, Cox DG, Angèle S, Romestaing P, Gérard JP, Hall J. Polymorphisms in the DNA repair gene XRCC1, breast cancer risk, and response to radiotherapy. Cancer Epidemiol Biomarkers Prev 2003;12:1168-74. 
|
| 16. | Hao B, Wang H, Zhou K, Li Y, Chen X, Zhou G, et al. Identification of genetic variants in base excision repair pathway and their associations with risk of esophageal squamous cell carcinoma. Cancer Res 2004;64:4378-84. 
|
| 17. | Wang C, Sun Y, Han R. XRCC1 genetic polymorphisms and bladder cancer susceptibility: A meta-analysis. Urology 2008;72:869-72. 
|
| 18. | Kiyohara C, Takayama K, Nakanishi Y. Association of genetic polymorphisms in the base excision repair pathway with lung cancer risk: A meta-analysis. Lung Cancer 2006;54:267-83. 
|
| 19. | Duell EJ, Millikan RC, Pittman GS, Winkel S, Lunn RM, Tse CK, et al. Polymorphisms in the DNA repair gene XRCC1 and breast cancer. Cancer Epidemiol Biomarkers Prev 2001;10:217-22. 
|
| 20. | Ratnasinghe D, Yao SX, Tangrea JA, Qiao YL, Andersen MR, Barrett MJ, et al. Polymorphisms of the DNA repair gene XRCC1 and lung cancer risk. Cancer Epidemiol Biomarkers Prev 2001;10:119-23. 
|
| 21. | Stern MC, Umbach DM, van Gils CH, Lunn RM, Taylor JA. DNA repair gene XRCC1 polymorphisms, smoking, and bladder cancer risk. Cancer Epidemiol Biomarkers Prev 2001;10:125-31. 
|
| 22. | Rossit AR, Cabral IR, Hackel C, da Silva R, Froes ND, Abdel-Rahman SZ. Polymorphisms in the DNA repair gene XRCC1 and susceptibility to alcoholic liver cirrhosis in older Southeastern Brazilians. Cancer Lett 2002;180:173-82. 
|
| 23. | Parildar-Karpuzoðlu H, Doðru-Abbasoðlu S, Hanagasi HA, Karadað B, Gürvit H, Emre M, et al . Single nucleotide polymorphisms in base-excision repair genes hOGG1, APE1 and XRCC1 do not alter risk of Alzheimer's disease. Neurosci Lett 2008;442:287-91. 
|
| 24. | Qian Y, Chen W, Wu J, Tao T, Bi L, Xu W, et al. Association of polymorphism of DNA repair gene XRCC1 with sporadic late-onset Alzheimer's disease and age of onset in elderly Han Chinese. J Neurol Sci 2010;295:62-5. 
|
| 25. | Zhao XH, Jia G, Liu YQ, Liu SW, Yan L, Jin Y, et al. Association between polymorphisms of DNA repair gene XRCC1 and DNA damage in asbestos-exposed workers. Biomed Environ Sci 2006;19:232-8. 
|
| 26. | Yousaf S, Khan MI, Micheal S, Akhtar F, Ali SH, Riaz M, et al. XRCC1 and XPD DNA repair gene polymorphisms: A potential risk factor for glaucoma in the Pakistani population. Mol Vis 2011;17:1153-63. 
|
| 27. | Güven M, Unal M, Batar B, Eroðlu E, Devarnoðlu K, Tamçelik N, et al . Polymorphisms of DNA repair genes XRCC1 and XPD and risk of primary open angle glaucoma (POAG). Mol Vis 2007;13:12-7. 
|
| 28. | Luo YF, Wang BB, Zhou Z, Ding XC, Hu SS, Zhou GK, et al. Polymorphisms of the DNA repair genes XPD and XRCC1 and the risk of age-related cataract development in Han Chinese. Curr Eye Res 2011;36:632-6. 
|
| 29. | Unal M, Güven M, Batar B, Ozaydin A, Sarici A, Devranoðlu K. Polymorphisms of DNA repair genes XPD and XRCC1 and risk of cataract development. Exp Eye Res 2007;85:328-34. 
|
| 30. | Padma G, Mamata M, Reddy KR, Padma T. Polymorphisms in two DNA repair genes (XPD and XRCC1) - Association with age related cataracts. Mol Vis 2011;17:127-33. 
|
| 31. | Attar R, Cacina C, Sozen S, Attar E, Agachan B. DNA repair genes in endometriosis. Genet Mol Res 2010;9:629-36. 
|
| 32. | Gu A, Ji G, Liang J, Xia Y, Lu N, Wu B, et al. DNA repair gene XRCC1 and XPD polymorphisms and the risk of idiopathic azoospermia in a Chinese population. Int J Mol Med 2007;20:743-7. 
|
| 33. | Yang SF, Xu YJ, Xie JG, Zhang Zx. hOGG1 Ser326Cys and XRCC1 Arg399Gln polymorphisms associated with chronic obstructive pulmonary disease. Chin Med J (Engl) 2009;122:960-6. 
|
| 34. | Guven M, Guven GS, Oz E, Ozaydin A, Batar B, Ulutin T, et al. DNA repair gene XRCC1 and XPD polymorphisms and their association with coronary artery disease risks and micronucleus frequency. Heart Vessels 2007;22:355-60. 
|
| 35. | Bazo AP, Salvadori D Jr, Salvadori RA, Sodré LP, da Silva GN, de Camargo EA, et al. DNA repair gene polymorphism is associated with the genetic basis of atherosclerotic coronary artery disease. Cardiovasc Pathol 2011;20:e9-15. 
|
| 36. | Sterpone S, Cornetta T, Angioni A, Fiscarelli E, Lucidi V, Testa A, et al. DNA damage and related modifier genes in Italian cystic fibrosis patients. Biol Res 2009;42:477-86. 
|
| 37. | Bau DT, Hsieh YY, Wan L, Wang RF, Liao CC, Lee CC, et al. Polymorphism of XRCC1 codon arg 399 Gln is associated with higher susceptibility to endometriosis. Chin J Physiol 2007;50:326-9. 
|
| 38. | Lin YJ, Wan L, Huang CM, Chen SY, Huang YC, Lai CH, et al. Polymorphisms in the DNA repair gene XRCC1 and associations with systemic lupus erythematosus risk in the Taiwanese Han Chinese population. Lupus 2009;18:1246-51. 
|
| 39. | Sobti RC, Mahdi SA, Berhane N, Hosseini SA, Kler R, Kuttiat V, et al. The influence of variations in the DNA repair (XRCC1) gene on HIV-1/AIDS among Indian population. Folia Biol (Praha) 2009;55:183-6. 
|
| 40. | Warcho³ T, Mostowska A, Lianeri M, L¹cki JK, Jagodziñski PP. XRCC1 Arg399Gln gene polymorphism and the risk of systemic lupus erythematosus in the Polish population. DNA Cell Biol 2012;31:50-6. 
|
| 41. | Görgün E, Güven M, Unal M, Batar B, Güven GS, Yenerel M, et al. Polymorphisms of the DNA repair genes XPD and XRCC1 and the risk of age-related macular degeneration. Invest Ophthalmol Vis Sci 2010;51:4732-7. 
|
| 42. | Vural P, Deðirmencioðlu S, Doðru-Abbasoðlu S, Saral NY, Akgül C, Uysal M. Genetic polymorphisms in DNA repair gene APE1, XRCC1 and XPD and the risk of pre-eclampsia. Eur J Obstet Gynecol Reprod Biol 2009;146:160-4. 
|
| 43. | Chiang CC, Tsai YY, Bau DT, Cheng YW, Tseng SH, Wang RF, et al. Pterygium and genetic polymorphisms of the DNA repair enzymes XRCC1, XPA, and XPD. Mol Vis 2010;16:698-704. 
|
| 44. | Chen PL, Yeh KT, Tsai YY, Koeh H, Liu YL, Lee H, et al. XRCC1, but not APE1 and hOGG1 gene polymorphisms is a risk factor for pterygium. Mol Vis 2010;16:991-6. 
|
| 45. | Ferguson HR, Wild CP, Anderson LA, Murphy SJ, Johnston BT, Murray LJ, et al. No association between hOGG1, XRCC1, and XPD polymorphisms and risk of reflux esophagitis, Barrett's esophagus, or esophageal adenocarcinoma: Results from the factors influencing the Barrett's adenocarcinoma relationship case-control study. Cancer Epidemiol Biomarkers Prev 2008;17:736-9. 
|
| 46. | Koyama A, Kubota Y, Shimamura T, Horiuchi S. Possible association of the X-ray cross complementing gene 1 (XRCC1) Arg280His polymorphism as a risk for rheumatoid arthritis. Rheumatol Int 2006;26:749-51. 
|
| 47. | Derakhshandeh S, Saadat I, Farrashbandi H, Saadat M. Association between genetic polymorphism of XRCC1 Arg194Trp and risk of schizophrenia. Psychiatry Res 2009;169:186. 
|
| 48. | Saadat M, Pakyari N, Farrashbandi H. Genetic polymorphism in the DNA repair gene XRCC1 and susceptibility to schizophrenia. Psychiatry Res 2008;157:241-5. 
|
| 49. | Olshan AF, Shaw GM, Millikan RC, Laurent C, Finnell RH. Polymorphisms in DNA repair genes as risk factors for spina bifida and orofacial clefts. Am J Med Genet A 2005;135:268-73. 
|
| 50. | Kasznicki J, Krupa R, B³asiak J, Drzewoski J. Association between polymorphisms of the DNA repair genes XRCC1 and hOGG1 and type 2 diabetes mellitus in the Polish population. Pol Arch Med Wewn 2009;119:122-8. 
|
| 51. | Batar B, Guven M, Onaran I, Tutluoglu B, Kanigur-Sultuybek G. DNA repair gene XRCC1 polymorphisms and the risk of asthma in a Turkish population. Allergy Asthma Proc 2010;31:349-54. 
|
| 52. | Xie J, Yang S, Xu Y, Zhang Z. XRCC1 Arg194Trp polymorphism and risk of chronic obstructive pulmonary disease. J Huazhong Univ Sci Technolog Med Sci 2009;29:551-6. 
|
| 53. | Ji G, Gu A, Zhu P, Xia Y, Zhou Y, Hu F, et al. Joint effects of XRCC1 polymorphisms and polycyclic aromatic hydrocarbons exposure on sperm DNA damage and male infertility. Toxicol Sci 2010;116:92-8. 
|
| 54. | Frank B, Müller H, Weck MN, Klopp N, Illig T, Raum E, et al. DNA repair gene polymorphisms and risk of chronic atrophic gastritis: A case-control study. BMC Cancer 2011;11:440. 
|
| 55. | Doðru-Abbasoðlu S, Aykaç-Toker G, Hanagasi HA, Gürvit H, Emre M, Uysal M. The Arg194Trp polymorphism in DNA repair gene XRCC1 and the risk for sporadic late-onset Alzheimer's disease. Neurol Sci 2007;28:31-4. 
|
| 56. | Bassi C, Xavier Dj, Palomino G, Nicolucci P, Soares C, Sakamoto-Hojo E, et al. Efficiency of the DNA repair and polymorphisms of the XRCC1, XRCC3 and XRCC4 DNA repair genes in systemic lupus erythematosus. Lupus 2008;17:988-95. 
|
| 57. | Ioannidis JP, Boffetta P, Little J, O'Brien TR, Uitterlinden AG, Vineis P, et al. Assessment of cumulative evidence on genetic associations: Interim guidelines. Int J Epidemiol 2008;37:120-32. 
|
| 58. | Liu HP, Lin WY, Wu BT, Liu SH, Wang WF, Tsai CH, et al. Evaluation of the poly(ADP-ribose) polymerase-1 gene variants in Alzheimer's disease. J Clin Lab Anal 2010;24:182-6. 
|
| 59. | Kleiman NJ, Wang RR, Spector A. Hydrogen peroxide-induced DNA damage in bovine lens epithelial cells. Mutat Res 1990;240:35-45. 
|
| 60. | Spector A. Oxidative stress-induced cataract: Mechanism of action. FASEB J 1995;9:1173-82. 
|
| 61. | Reddy VN, Giblin FJ, Lin LR, Chakrapani B. The effect of aqueous humor ascorbate on ultraviolet-B-induced DNA damage in lens epithelium. Invest Ophthalmol Vis Sci 1998;39:344-50. 
|
| 62. | Risa Ø, Saether O, Löfgren S, Söderberg PG, Krane J, Midelfart A. Metabolic changes in rat lens after in vivo exposure to ultraviolet irradiation: Measurements by high resolution MAS 1H NMR spectroscopy. Invest Ophthalmol Vis Sci 2004;45:1916-21. 
|
| 63. | Pendergrass W, Penn P, Possin D, Wolf N. Accumulation of DNA, nuclear and mitochondrial debris, and ROS at sites of age-related cortical cataract in mice. Invest Ophthalmol Vis Sci 2005;46:4661-70. 
|
| 64. | McCarty CA, Taylor HR. A review of the epidemiologic evidence linking ultraviolet radiation and cataracts. Dev Ophthalmol 2002;35:21-31. 
|
| 65. | Capellá G, Pera G, Sala N, Agudo A, Rico F, Del Giudicce G, et al. DNA repair polymorphisms and the risk of stomach adenocarcinoma and severe chronic gastritis in the EPIC-EURGAST study. Int J Epidemiol 2008;37:1316-25. 
|
| 66. | Andreassi MG. Coronary atherosclerosis and somatic mutations: An overview of the contributive factors for oxidative DNA damage. Mutat Res 2003;543:67-86. 
|
| 67. | Dinçer Y, Akçay T, Ilkova H, Alademir Z, Ozbay G. DNA damage and antioxidant defense in peripheral leukocytes of patients with Type I diabetes mellitus. Mutat Res 2003;527:49-55. 
|
| 68. | Duell EJ, Wiencke JK, Cheng TJ, Varkonyi A, Zuo ZF, Ashok TD, et al. Polymorphisms in the DNA repair genes XRCC1 and ERCC2 and biomarkers of DNA damage in human blood mononuclear cells. Carcinogenesis 2000;21:965-71. 
|
| 69. | Tuimala J, Szekely G, Gundy S, Hirvonen A, Norppa H. Genetic polymorphisms of DNA repair and xenobiotic-metabolizing enzymes: Role in mutagen sensitivity. Carcinogenesis 2002;23:1003-8. 
|
| 70. | Wang Y, Spitz MR, Zhu Y, Dong Q, Shete S, Wu X. From genotype to phenotype: Correlating XRCC1 polymorphisms with mutagen sensitivity. DNA Repair (Amst) 2003;2:901-8. 
|
| 71. | Patel AV, Calle EE, Pavluck AL, Feigelson HS, Thun MJ, Rodriguez C. A prospective study of XRCC1 (X-ray cross-complementing group 1) polymorphisms and breast cancer risk. Breast Cancer Res 2005;7:R1168-73. 
|
| 72. | Lao T, Gu W, Huang Q. A meta-analysis on XRCC1 R399Q and R194W polymorphisms, smoking and bladder cancer risk. Mutagenesis 2008;23:523-32. 
|
| 73. | Huang Y, Li L, Yu L. XRCC1 Arg399Gln, Arg194Trp and Arg280His polymorphisms in breast cancer risk: A meta-analysis. Mutagenesis 2009;24:331-9. 
|
| 74. | Yan L, Yanan D, Donglan S, Na W, Rongmiao Z, Zhifeng C. Polymorphisms of XRCC1 gene and risk of gastric cardiac adenocarcinoma. Dis Esophagus 2009;22:396-401. 
|
| 75. | Fard-Esfahani P, Fard-Esfahani A, Fayaz S, Ghanbarzadeh B, Saidi P, Mohabati R, et al. Association of Arg194Trp, Arg280His and Arg399Gln polymorphisms in X-ray repair cross-complementing group 1 gene and risk of differentiated thyroid carcinoma in Iran. Iran Biomed J 2011;15:73-8. 
|
| 76. | Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev 2002;11:1513-30. 
|
| 77. | Hung RJ, Hall J, Brennan P, Boffetta P. Genetic polymorphisms in the base excision repair pathway and cancer risk: A HuGE review. Am J Epidemiol 2005;162:925-42. 
|
| 78. | Qu T, Morimoto K. X-ray repair cross-complementing group 1 polymorphisms and cancer risks in Asian populations: A mini review. Cancer Detect Prev 2005;29:215-20. 
|
| 79. | Hu Z, Ma H, Chen F, Wei Q, Shen H. XRCC1 polymorphisms and cancer risk: A meta-analysis of 38 case-control studies. Cancer Epidemiol Biomarkers Prev 2005;14:1810-8. 
|
| 80. | Lunn RM, Langlois RG, Hsieh LL, Thompson CL, Bell DA. XRCC1 polymorphisms: Effects on aflatoxin B1-DNA adducts and glycophorin A variant frequency. Cancer Res 1999; 59:2557-61. 
|
[Figure 1], [Figure 2], [Figure 3], [Figure 4], [Figure 5], [Figure 6], [Figure 7], [Figure 8], [Figure 9], [Figure 10], [Figure 11], [Figure 12]
[Table 1], [Table 2], [Table 3], [Table 4], [Table 5], [Table 6]
|






