|
 
 |
|
ORIGINAL ARTICLE |
|
|
|
| Year : 2014 | Volume
: 20
| Issue : 2 | Page : 142-147 |
| |
Methylenetetrahydrofolate reductase polymorphism is not risk factor for Down syndrome in North India
Vandana Rai, Upendra Yadav, Pradeep Kumar, Sushil Kumar Yadav
Department of Biotechnology, Human Molecular Genetics Laboratory, VBS Purvanchal University, Jaunpur, Uttar Pradesh, India
| Date of Web Publication | 14-Oct-2014 |
Correspondence Address:
Vandana Rai
Department of Biotechnology, Human Molecular Genetics Laboratory, VBS Purvanchal University, Jaunpur - 222 001, Uttar Pradesh
India
 Source of Support: This study was supported by the financial assistance from Department of Biotechnology, New Delhi (No BT/PR98887/SPD/11/1028/2007), for providing the financial support to Vandana Rai to conduct this study,, Conflict of Interest: None
DOI: 10.4103/0971-6866.142858

 Abstract Abstract | | |
Background: Down syndrome (DS) is the most common cause of mental retardation of genetic etiology with the prevalence rate of 1/700 to 1/1000 live births worldwide. Several polymorphisms in folate/homocysteine metabolism pathways genes have been reported as a risk factor in women for bearing DS child, but very few studies investigated these polymorphisms in DS cases whether there are a risk factor for being DS or not.
Objective: We have investigated the association of methylenetetrahydrofolate reductase (MTHFR) with the occurrence of DS in Indian population. MTHFR is one of the key regulatory enzymes involved in the metabolic pathway of homocysteine responsible for the reduction of methyltetrahydrofolate. A total of 32 DS cases and 64 age, sex matched controls were genotyped for MTHFR C677T polymorphism by polymerase chain reaction-restriction fragment length polymorphism.
Results: The observed genotype frequencies were CC = 0.81; CT = 0.17 and TT = 0.02 in controls and CC = 0.81 and CT = 0.19 in DS cases. Frequency of T allele in DS and controls were 0.09 and 0.1, respectively. Significant difference in the distribution of mutant 677T allele was not observed between DS cases and controls (odds ratio = 0.915; 95% confidence intervals: 0.331-2.53; P = 0.864).
Conclusion: Results of this study indicate that MTHFR C677T polymorphism is not risk factor for DS.
Keywords: Down syndrome, homocysteine, methylation, methylenetetrahydrofolate reductase, polymorphism
How to cite this article:
Rai V, Yadav U, Kumar P, Yadav SK. Methylenetetrahydrofolate reductase polymorphism is not risk factor for Down syndrome in North India. Indian J Hum Genet 2014;20:142-7 |
How to cite this URL:
Rai V, Yadav U, Kumar P, Yadav SK. Methylenetetrahydrofolate reductase polymorphism is not risk factor for Down syndrome in North India. Indian J Hum Genet [serial online] 2014 [cited 2016 Aug 23];20:142-7. Available from: http://www.ijhg.com/text.asp?2014/20/2/142/142858 |
 Introduction Introduction | |  |
Down syndrome (DS) (MIM 190685), is the most frequent genetic cause of mental retardation, resulting from the presence of three copies of genes located on chromosome 21 and the most common chromosomal disorder in newborns with prevalence of 1/700 live births. Advanced maternal age is the only well-documented risk factor for nondisjunction of chromosome 21. [1] In spite of high prevalence, the cellular and molecular mechanisms underlying meiotic nondisjunction are not yet understood, and much research have been done on factors that influence the rate of chromosome nondisjunction. The relationship between chromosomal nondisjunction and folate metabolism has drawn attention in the past decade. Folate and methylenetetrahydrofolate reductase (MTHFR) are both involved in many complex biochemical reactions. A deficiency in cellular folates and methyl-donors may be associated with abnormal DNA methylation, defective chromosome recombination, and abnormal chromosome segregation. [2] James et al. [3] were the first to propose the hypothesis that altered DNA methylation patterns resulting from abnormal folate metabolism may increase DNA hypomethylation in centromeric DNA. Several published papers emphasized that folate metabolism is modulated by genetic factor like single nucleotide polymorphisms (SNPs) in MTHFR, methionine synthase (MTR) and MTR reductase (MTRR) genes and dietary folate and B vitamin supplements. MTHFR is the most studied enzyme and responsible for the reduction of methylenetetrahydrofolate, which is a key single-carbon donor that takes part in nucleotide synthesis; remethylation of homocysteine to methionine; S-adenosyl-methionine (SAM) synthesis; and the methylation of DNA, proteins, neurotransmitters, and phospholipids. Reduced MTHFR activity results in an increased requirement for folic acid to maintain normal homocysteine remethylation to methionine. In the absence of sufficient folic acid and/or dysfunctional MTHFR, intracellular homocysteine accumulates, methionine synthesis is reduced, and methylation reactions are interrupted. Increased homocysteine and decreased methionine cause decreased SAM to S-adenosylhomocysteine ratio, which takes part in DNA methylation. [4]
The MTHFR gene (OMIM 607093) has been mapped at short arm of chromosome 1 (1p36.3) and more than 40 polymorphism have been described in MTHFR, but C677T (rs1801133) is the most common [5] in which a cytosine is replaced by thymine at 677 th position (alanine →valine in protein). C677T mutation was shown to render the enzyme thermolabile and homozygotes (T/T) and heterozygotes (C/T) had about a 70% and 35% reduced MTHFR enzymatic activity. Frequency of mutant T allele differs in various ethnic and geographical populations of the world. For example, the allele frequency ranges from 0.20 to 0.55 in Europeans and from 0.04 to 0.38 in Asian populations. [6],[7],[8],[9],[10],[11] C677T polymorphism has been reported as a risk factor for several diseases and/or clinical conditions including neural tube defects (NTD), [12] orofacial clefts, [13] cardiovascular diseases [5] and cancer. [14] Several studies have been published that aim to evaluate the role of maternal MTHFR C677T polymorphism as a risk factor for DS pregnancy [4],[15],[16],[17],[18],[19],[20],[21] but very few studies are carried out to evaluate MTHFR C677T polymorphism in DS cases as a risk factor. Thus, the objective of this study is to analyze C677T polymorphism in DS cases and control group.
 Materials and Methods Materials and Methods | |  |
The present study was approved by the Institutional Ethics Committee of VBS Purvanchal University, Jaunpur. Informed written consent was taken from each subject after fully verbal explanation of the nature of the study. In the present case-control study, a total of 32 DS cases and 64 age, sex matched controls from eastern Uttar Pradesh were recruited. Genomic DNA was extracted from peripheral blood of all the subjects using standard protocol of Bartlett and White. [22] Genotyping for the MTHFR point mutation C677T was carried out by polymerase chain reaction (PCR)-restriction fragment length polymorphism method of Frosst et al. [5] PCR was carried out in a total reaction volume of 15 μl consisting of 1.5 μl of PCR buffer (×10), 1.5 μl of dNTPs mix (2.5 mM), 4 pM of each of the forward (5'TGAAGGAGAAGGTGTCTGCGGGA-3') and reverse (5'AGGACGGTGCGGTGAGAGTG-3') primers, 1.5 units of taq DNA polymerase enzyme (Genei) and 200 ng genomic DNA. PCR was carried out in MJ thermal cycler (BioRad, USA). PCR amplification conditions included initial denaturation at 94°C for 4 min followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 65°C for 1 min and polymerization at 72°C for 1 min and a final polymerization at 72°C for 10 min. The amplicons were digested with HinfI as the C677T mutation creates a restriction site for it, and resolved in a 3% agarose gel.
Statistical analysis
Deviation from Hardy-Weinberg equilibrium was determined by Chi-square test. Association between MTHFR C677T polymorphism and DS risk was calculated as odds ratio (OR) estimates with 95% confidence intervals (95% CI). Logistic regression analyses considering three model for effect of polymorphism were performed: Co-dominant (TT vs. CC and CT vs. CC), dominant (TT and CT vs. CC) and recessive (TT vs. CT and CC). Association was considered significant at P < 0.05. All statistical analyses were performed by online program available at ihg.gsf.de/cgi-bin/hw/hwa1.pl .
 Results and Discussion Results and Discussion | |  |
The DNA from the controls and DS cases is PCR amplified and HinfI-digested amplicon is resolved in 3% agarose gel. The wild type allele C is not cut by the enzyme and yields a 198 bp fragment after amplification. The T allele, on the contrary, creates a site for HinfI leading to its digestion and producing two bands of 175 and 23 bp. As the 23 bp fragment is not resolvable in the gel, occurrence of the 175 bp fragment is taken to indicate the presence of T allele. [Figure 1] illustrates the genotypes (CC and CT) in DS cases. We performed two different types of statistical analyses: First, a comparison of allele frequencies between case and control individuals (C and T alleles); and second, a comparison of the frequencies of the three genotypes (CC, CT, and TT genotypes) among case and control individuals. A summary of allele and genotype frequencies of MTHFR C677T polymorphism in DS cases and controls given in [Table 1]. The frequency of mutant MTHFR T allele was 0.09 in DS cases and 0.1 in controls. Genotype frequencies in controls were 0.81, 0.17, and 0.02 for CC, CT and TT, respectively and 0.81 and 0.19 for CC and CT, respectively in DS cases. TT genotype was not observed in any DS case. Analyses of allele and genotype frequencies of MTHFR C677T polymorphism have revealed nonsignificant difference between cases and controls. As is evident from the [Table 1], while C is the dominant allele in the controls as well as the cases, the proportion of T allele in the DS cases (9%) is nearly similar to the proportion of T allele in the controls (10%). The OR for T allele in cases against controls has been calculated (OR = 0.915; 95% CI: 0.331-2.53; P = 0.864) and showed no association between T allele and DS. CC genotype percentage (81.25) was similar in both controls as well as DS cases, whereas against 17.19% (11/64) CT in controls, there were 18.75% CT (6/32) in the DS cases. The heterozygous genotype differences were observed between case and controls; therefore, OR has been also calculated for CT versus CC genotypes between DS cases and controls (OR = 1.091; 95% CI: 0.36-3.2; P = 0.876). Although TT genotype was not observed in case samples, but we calculated OR for TT versus CC (OR = 0.66; 95% CI: 0.26-16.76; P = 0.480). Further OR for risk genotypes TT and CT versus CC has been also calculated (OR = 1.00; 95% CI: 0.337-2.966; P = 1.0) [Table 2].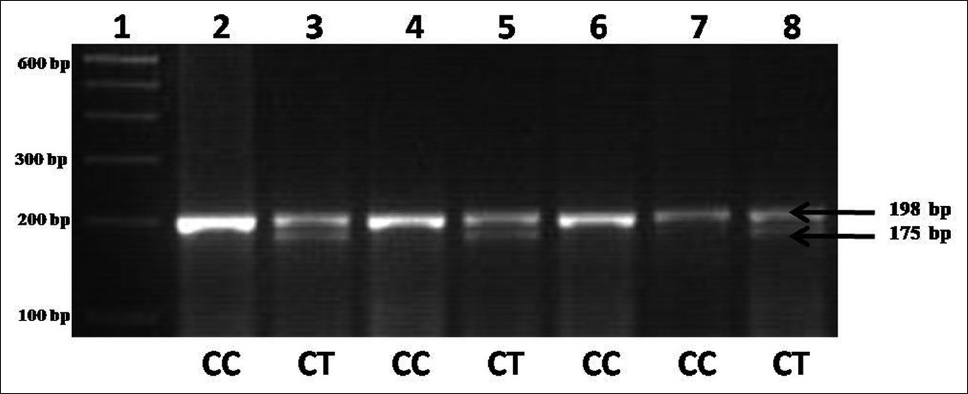 | Figure 1: Agarose gel picture showing Hinf I digested amplicons showing genotypes of methylenetetrahydrofolate reductase
Click here to view |
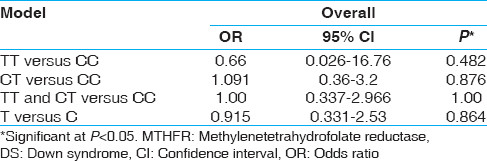 | Table 2: Association between MTHFR C677T genotype and DS cases (n=32) and controls (n=64)
Click here to view |
Several studies performed on human cell cultures, in vivo studies in humans and studies involving animal models have demonstrated that folate depletion from the media, or inadequate folate dietary intake, result in DNA hypomethylation, chromosome breakage, and aneuploidy. [23] For this reason, it has been postulated that impairments in folate and homocysteine metabolism due to genetic polymorphisms of folate metabolic enzymes could predispose women to abnormal chromosome segregation and act as risk factors for a DS pregnancy. [3] Since 1999, several epidemiological studies have been performed on mother of DS, aimed at evaluating the role of folate and homocysteine pathway gene polymorphisms in the risk of a DS offspring and several associations have been observed. [3],[4],[17],[24],[25] MTHFR C677T variant due to less enzymatic activity leads to hypomethylation of centromeric DNA, which may be the major cause of missegregation of chromosomes during meiosis and results in trisomy 21. [4] Hassold et al. [26] analyzed polymorphism of MTHFR and MTRR maternal genes in trisomy of several chromosomes and compared the distribution of genotypes to those of control populations and observed a significant increase in the MTHFR C677T polymorphism in mothers of trisomy cases. Several population-based studies and experiments with animal models have shown that periconceptional multivitamin/folic acid supplementation has a strong protective effect during early stages of embryo development, resulting in a significant reduction in the occurrence of development defects, including NTD, congenital heart defects, limb defects, and orofacial clefts. [27] Low or inadequate intake of folic acid are involved in the disruption of methionine metabolism, because methylenetetrahydrofolate, the primary form of folate in the circulation, acts as the carbon donor for homocysteine remethylation to yield methionine and tetrahydrofolate. [28] Therefore, genes that participate in homocysteine/folate metabolism are attractive candidates for investigation of the mechanisms underlying the protective role of folic acid in birth defects and DS.
The impact of folic acid intake on pregnancy outcome, however, is modified by polymorphisms in both the maternal and fetal genes that code for enzymes involved in folate metabolism. [27] Data on the genotype and allele frequencies of MTHFR C677T polymorphism in DS mothers compared to non-DS mothers are numerous and show significant differences, but reports regarding distribution of MTHFR mutant allele and genotype frequencies in DS individuals are very few. [19],[29],[30],[31],[32],[33] Several clinical and experimental studies have hypothesized/reported that patients with DS have disturbed one-carbon metabolism. [2],[3] t-Hcy, vitamin B-12 and folate are metabonutritional factors directly related to this metabolism. These factors along with the associated genetic polymorphisms might aggravate the age related mental retardation in DS.
Guéant et al. [34] have reported that carriers of the MTHFR 677T allele DS individuals had a lower IQ than those carrying the corresponding wild genotype. The influence of MTHFR 677T allele may correspond to decreased remethylation of homocysteine due to reduced activity of MTHFR, eventually accentuated by reduced availability of vitamin B12 cofactor of MTR. None of the other genetic polymorphisms of the one-carbon metabolism influenced the level of mental retardation in DS. High homocysteine concentration impairs DNA repair in hippocampal neurons, promotes apoptosis, hypersensitivity to exitotoxicity and oxidative stress, and it potentiates the neurone toxicity of beta-amyloid peptide, the proteolytic product of amyloid precursor protein. [35] In addition to the over expression of cystathionine beta synthase (CβS) due to three copies of CβS genes in DS cases, another phenotypic abnormality related to chromosome 21 genes is the over expression of amyloid precursor protein, which confers a predominant role to the pathway of amyloid precursor protein in the neurodegeneration in DS dementia. [36]
This study failed to reveal any association between C677T polymorphism and risk of being DS child, but we conclude that this SNP might be responsible for low IQ, severe mental retardation, dementia, neurodegeneration and other DS symptoms in severe conditions in comparison to CC DS. DS individuals with TT genotype might be particularly sensitive to the status of folate and several B vitamins and should be supplemented for personalized nutritional recommendations especially for folate, B-6 and B-12 vitamins. In conclusion, as there was no statistical difference in genotype distribution between cases and controls, we suggest that the analysis of MTHFR genotyping for C677T polymorphism alone need not be considered to find out whether there is a genetic risk for development of DS, polymorphisms of other folate/homocysteine pathway genes like CβS and replication factor C, which are located on chromosome 21 should be studied as a risk factor along with the MTHFR gene.
 Acknowledgments Acknowledgments | |  |
The authors are grateful to all patients, their families, and control volunteers for their participation in the study. This study was supported by the financial assistance from Department of Biotechnology, New Delhi (No BT/PR98887/SPD/11/1028/2007), for providing the financial support to Vandana Rai to conduct this study.
 References References | |  |
| 1. | Penrose LS. The relative effects of paternal and maternal age in mongolism. 1933. J Genet 2009;88:9-14.  [ PUBMED] |
| 2. | Pogribna M, Melnyk S, Pogribny I, Chango A, Yi P, James SJ. Homocysteine metabolism in children with Down syndrome: In vitro modulation. Am J Hum Genet 2001;69:88-95.  |
| 3. | James SJ, Pogribna M, Pogribny IP, Melnyk S, Hine RJ, Gibson JB, et al. Abnormal folate metabolism and mutation in the methylenetetrahydrofolate reductase gene may be maternal risk factors for Down syndrome. Am J Clin Nutr 1999;70:495-501.  |
| 4. | Hobbs CA, Sherman SL, Yi P, Hopkins SE, Torfs CP, Hine RJ, et al. Polymorphisms in genes involved in folate metabolism as maternal risk factors for Down syndrome. Am J Hum Genet 2000;67:623-30.  |
| 5. | Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG, et al. A candidate genetic risk factor for vascular disease: A common mutation in methylenetetrahydrofolate reductase. Nat Genet 1995;10:111-3.  [ PUBMED] |
| 6. | Schneider JA, Rees DC, Liu YT, Clegg JB. Worldwide distribution of a common methylenetetrahydrofolate reductase mutation. Am J Hum Genet 1998;62:1258-60.  [ PUBMED] |
| 7. | Rady PL, Szucs S, Grady J, Hudnall SD, Kellner LH, Nitowsky H, et al. Genetic polymorphisms of methylenetetrahydrofolate reductase (MTHFR) and methionine synthase reductase (MTRR) in ethnic populations in Texas; A report of a novel MTHFR polymorphic site, G1793A. Am J Med Genet 2002;107:162-8.  |
| 8. | Wilcken B, Bamforth F, Li Z, Zhu H, Ritvanen A, Renlund M, et al. Geographical and ethnic variation of the 677C>T allele of 5,10 methylenetetrahydrofolate reductase (MTHFR): Findings from over 7000 newborns from 16 areas worldwide. J Med Genet 2003;40:619-25.  [ PUBMED] |
| 9. | Erdogan MO, Yildiz SH, Solak M, Eser O, Cosar E, Eser B, et al. C677T polymorphism of the methylenetetrahydrofolate reductase gene does not affect folic acid, vitamin B12, and homocysteine serum levels in Turkish children with neural tube defects. Genet Mol Res 2010;9:1197-203.  |
| 10. | Rai V, Yadav U, Kumar P, Yadav SK. Methylenetetrahydrofolate reductase polymorphism (c677t) in Muslim population of eastern Uttar Pradesh, India. Indian J Med Sci 2010;64:219-23.  [ PUBMED]  |
| 11. | Rai V, Yadav U, Kumar P. Prevalence of methylenetetrahydrofolate reductase C677T polymorphism in eastern Uttar Pradesh. Indian J Hum Genet 2012;18:43-6.  [ PUBMED]  |
| 12. | Godbole K, Gayathri P, Ghule S, Sasirekha BV, Kanitkar-Damle A, Memane N, et al. Maternal one-carbon metabolism, MTHFR and TCN2 genotypes and neural tube defects in India. Birth Defects Res A Clin Mol Teratol 2011;91:848-56.  |
| 13. | Blanton SH, Henry RR, Yuan Q, Mulliken JB, Stal S, Finnell RH, et al. Folate pathway and nonsyndromic cleft lip and palate. Birth Defects Res A Clin Mol Teratol 2011;91:50-60.  |
| 14. | Kotsopoulos J, Zhang WW, Zhang S, McCready D, Trudeau M, Zhang P, et al. Polymorphisms in folate metabolizing enzymes and transport proteins and the risk of breast cancer. Breast Cancer Res Treat 2008;112:585-93.  |
| 15. | Boduroğlu K, Alanay Y, Koldan B, Tunçbilek E. Methylenetetrahydrofolate reductase enzyme polymorphisms as maternal risk for Down syndrome among Turkish women. Am J Med Genet A 2004;127A:5-10.  |
| 16. | da Silva LR, Vergani N, Galdieri Lde C, Ribeiro Porto MP, Longhitano SB, Brunoni D, et al. Relationship between polymorphisms in genes involved in homocysteine metabolism and maternal risk for Down syndrome in Brazil. Am J Med Genet A 2005;135:263-7.  |
| 17. | Coppedè F, Marini G, Bargagna S, Stuppia L, Minichilli F, Fontana I, et al. Folate gene polymorphisms and the risk of Down syndrome pregnancies in young Italian women. Am J Med Genet A 2006;140:1083-91.  |
| 18. | Martínez-Frías ML, Pérez B, Desviat LR, Castro M, Leal F, Rodríguez L, et al. Maternal polymorphisms 677C-T and 1298A-C of MTHFR, and 66A-G MTRR genes: Is there any relationship between polymorphisms of the folate pathway, maternal homocysteine levels, and the risk for having a child with Down syndrome? Am J Med Genet A 2006;140:987-97.  |
| 19. | Biselli JM, Goloni-Bertollo EM, Zampieri BL, Haddad R, Eberlin MN, Pavarino-Bertelli EC. Genetic polymorphisms involved in folate metabolism and elevated plasma concentrations of homocysteine: Maternal risk factors for Down syndrome in Brazil. Genet Mol Res 2008;7:33-42.  |
| 20. | Wang SS, Qiao FY, Feng L, Lv JJ. Polymorphisms in genes involved in folate metabolism as maternal risk factors for Down syndrome in China. J Zhejiang Univ Sci B 2008;9:93-9.  |
| 21. | Brandalize AP, Bandinelli E, dos Santos PA, Roisenberg I, Schüler-Faccini L. Evaluation of C677T and A1298C polymorphisms of the MTHFR gene as maternal risk factors for Down syndrome and congenital heart defects. Am J Med Genet A 2009;149A:2080-7.  |
| 22. | Bartlett JM, White A. Extraction of DNA from blood. In: Bartlett JM, Stirling D, editors. Methods in Molecular Biology, PCR Protocols. 2 nd ed. Totowa, NJ: Humana Press Inc.; 2003.  |
| 23. | Fenech M. The role of folic acid and Vitamin B12 in genomic stability of human cells. Mutat Res 2001;475:57-67.  [ PUBMED] |
| 24. | O'Leary VB, Parle-McDermott A, Molloy AM, Kirke PN, Johnson Z, Conley M, et al. MTRR and MTHFR polymorphism: Link to Down syndrome? Am J Med Genet 2002;107:151-5.  |
| 25. | Acácio GL, Barini R, Bertuzzo CS, Couto EC, Annichino-Bizzacchi JM, Júnior WP. Methylenetetrahydrofolate reductase gene polymorphisms and their association with trisomy 21. Prenat Diagn 2005;25:1196-9.  |
| 26. | Hassold TJ, Burrage LC, Chan ER, Judis LM, Schwartz S, James SJ, et al. Maternal folate polymorphisms and the etiology of human nondisjunction. Am J Hum Genet 2001;69:434-9.  |
| 27. | Botto LD, Yang Q. 5, 10-Methylenetetrahydrofolate reductase gene variants and congenital anomalies: A huge review. Am J Epidemiol 2000;151:862-77.  |
| 28. | Elmore CL, Wu X, Leclerc D, Watson ED, Bottiglieri T, Krupenko NI, et al. Metabolic derangement of methionine and folate metabolism in mice deficient in methionine synthase reductase. Mol Genet Metab 2007;91:85-97.  |
| 29. | Božovic IB, Vranekovic J, Cizmarevic NS, Mahulja-Stamenkovic V, Prpic I, Brajenovic-Milic B. MTHFR C677T and A1298C polymorphisms as a risk factor for congenital heart defects in Down syndrome. Pediatr Int 2011;53:546-50.  |
| 30. | Fillon-Emery N, Chango A, Mircher C, Barbé F, Bléhaut H, Herbeth B, et al. Homocysteine concentrations in adults with trisomy 21: Effect of B vitamins and genetic polymorphisms. Am J Clin Nutr 2004;80:1551-7.  |
| 31. | Bosco P, Guéant-Rodriguez RM, Anello G, Barone C, Namour F, Caraci F, et al. Methionine synthase (MTR) 2756 (A --> G) polymorphism, double heterozygosity methionine synthase 2756 AG/methionine synthase reductase (MTRR) 66 AG, and elevated homocysteinemia are three risk factors for having a child with Down syndrome. Am J Med Genet A 2003;121A:219-24.  |
| 32. | Amorim MR, Zanrosso CW, Magalhães IQ, Pereira SC, Figueiredo A, Emerenciano M, et al. MTHFR 677C-->T and 1298A-->C polymorphisms in children with Down syndrome and acute myeloid leukemia in Brazil. Pediatr Hematol Oncol 2008;25:744-50.  |
| 33. | Kuz'mina NS, Ushenkova LI, Shagirova ZhM, Sheikhaev GO, Mikhailov VF, Kurbatova LA, et al. Gene polymorphisms in patients with Down's syndrome. Zh Nevrol Psikhiatr Im S S Korsakova 2009;109:50-4.  |
| 34. | Guéant JL, Anello G, Bosco P, Guéant-Rodríguez RM, Romano A, Barone C, et al. Homocysteine and related genetic polymorphisms in Down's syndrome IQ. J Neurol Neurosurg Psychiatry 2005;76:706-9.  |
| 35. | Mattson MP, Shea TB. Folate and homocysteine metabolism in neural plasticity and neurodegenerative disorders. Trends Neurosci 2003;26:137-46.  |
| 36. | Hyman BT, West HL, Rebeck GW, Lai F, Mann DM. Neuropathological changes in Down's syndrome hippocampal formation. Effect of age and apolipoprotein E genotype. Arch Neurol 1995;52:373-8.  |
[Figure 1]
[Table 1], [Table 2]
|






