Comparing Solid Tumors with Cell Lines: Implications for Identifying Drug Resistance Genes in Cancer
Every year, more than a million new cases of cancer are diagnosed in the US. Regrettably, despite recent developments, effectiveness of chemotherapy is still rather limited for most types of cancer, including tumors of the colon, lung, kidney, pancreas, and liver. Why some cancers respond better than others may be explained by factors relating to the anatomy and physiology of the cancer- ridden organ or the pharmacokinetics of the drugs used to combat the disease. In addition, cancers may become resistant through cellular mechanisms, the prime example of which is the elevated expression of an ATP-dependent drug-efflux pump, termed P-glycoprotein (P-gp) (1). P-gp, also known as multidrug resistance protein1 (MDR1), is an integral membrane protein that transports a wide variety of chemotherapeutic molecules across cell membranes, thus keeping drug levels below a cell-killing threshold. The expression of MDR1 often increases during the course of treatment, and can hamper the chemotherapy of cancers originating from various tissues. That other cellular mechanisms, associated with the specific tissue of origin and the transformation process, must play additional roles is suggested by the observation that the tractability (i.e., responsiveness to chemotherapy) of a tumor extends to its disseminated metastases (2). That is, metasta-ses of tumors are as sensitive or refractory to treatment as are the original tumors.
To catalog possible cellular mechanisms of drug resistance, specimens obtained from solid tumors or metastases may be characterized on the basis of their expression profile as determined by microarray experiments. Current evidence suggests that cancer cells retain molecular signatures characteristic of their tissue of origin. Does this mean that gene expression patterns uniquely found in pancreatic cancers hold the key to understanding the intractability (i.e., resistance to chemotherapy) of that tumor type? Stein and colleagues sought to answer this puzzling question by comparing the genetic background of several tractable and intractable cancers (3). Focusing on six tissues known to give rise to cancers with increasing response to therapy, they mined the Serial Analysis of Gene Expression (SAGE) database to identify genes that were increasingly over- or under-expressed in solid tumors along the “axis of tractability” (Box 1) (Figure 1⇓). This approach, measuring the correlation between the levels of gene expression in a particular tissue and the tractability of tumors of that tissue, held the promise of distinguishing tissue-specific signatures that are unrelated to therapeutic response from those causally linked genes that determine therapeutic sensitivity. The resulting list revealed genetic themes relating to protein synthesis and intercellular inter-action. Specifically, genes essential to protein biosynthesis (and ribosome biogenesis) that are involved in proliferation are coordinately regulated and are significantly overexpressed in tumors associated with better patient survival, whereas genes encoding constituents of the extracellular matrix (ECM) that are involved in cell adhesion tend to be highly expressed in cancers with poor prognosis.
The Axis of Tractability: Classification of Tumors Based on Their Sensitivity to Currently Used Chemotherapies.
Stein defined the position of each tumor type on the axis of tractability based on a quantitative assessment of therapy-response. By mining the SEER [Surveillance, Epidemiology, and End Results (Program)] database (http://seer.cancer.gov/csr/1973_1999), which contains the five-year survival rates associated with tumors arising from different tissues, Stein found tumors of the pancreas, liver, colon, and lung to be the most intractable. Data suggested tumors of the testis to be the most tractable, whereas cancers of breast, ovary, and prostate origin occupied an intermediate position.
Drug-sensitivity is typically studied using cancer cell lines derived from various tissues. It is interesting to note that, compared to their corresponding solid tumors, cell lines become more sensitive to cytotoxic drugs, and this changed pattern of response does not strictly depend on the tissue of origin (4). Because in vitro conditions by no means resemble the in vivo environment, this increased sensitivity might be explained by external factors, such as the composition of the medium, lack of growth factors, temperature, etc. At the same time, gene expression patterns of the cells selected for in vitro propagation are likely to be different from that of the original tumor’s signature, and the differentially expressed genes may be responsible for the attenuated resistance. However, as has been shown for sixty cancer cell lines of various origins, expression patterns remain remarkably intact (as compared to the gene expression patterns in the original solid tumors) as it is normally a straightforward task to cluster cell lines by tissue of origin (5). To find the genetic basis for the difference between the sensitivity of cell lines and insensitivity of solid tumors, Stein focused his attention on genes whose expression levels were systematically changed through all tissue types. Strikingly, themes emerging from the list of differentially expressed genes were similar to those related to the tractability of the different cancer types. In other words, genes that are differentially expressed in solid tumors as compared to expression in their more drug-sensitive derivative cell lines predict general tractability of cancer.
Surprising as it is, the reoccurring themes of proliferation and adhesion as key determinants of tractability and chemosensitivity make sense. Survival of cells in the context of the tissue requires intercellular communication, a function less needed in a tissue culture flask. At the same time, in vitro cultured cells are selected for rapid growth, giving rise to the overexpression of genes involved in protein synthesis. Rapid growth of cells will, of course, result in a cancer that is more aggressive, but perhaps at the same time more susceptible to chemotherapy. This scenario is seen in hematological malignancies [e.g., fast-growing acute myeloid leukemia (AML) vs indolent, chronic myelogenous leukemia (CML)]. Conversely, increased cell adhesion has been widely accepted to result in decreased drug sensitivity, as exemplified by studies using cells grown as solid three-dimensional structures (i.e., multicellular tumor spheroids). To summarize, the data collected by Stein et al. suggest that: 1) solid tumors are adherent and inert as compared to their derivatives kept in tissue culture flasks, and 2) these characteristics increase with the intractability of the tumor types.
Candidate drugs for anticancer treatment are typically identified in in vitro models. A panel of sixty human cancer cell lines (the NCI-60) is routinely used by the Developmental Therapeutics program (DTP) of the National Cancer Institute (NCI) to screen novel chemical compounds. Although the sixty cell lines show an increased overall sensitivity to chemical compounds as compared to sensitivity in primary tumors, individual differences remain measurable. A sophisticated analysis, building on the correlation of gene expression profiles to patterns of chemosensitivity, has the potential to identify singular mechanisms that shape cellular response to treatment (6). As we have seen, the sensitivity pattern of the cells may not reflect tractability of the original tumor types, suggesting that the NCI60 screen might be exploring only a part of a tumor’s sensitivity to a potential drug. Thus, genes identified by Stein and coworkers should be included as potential targets of improved anticancer reagents.
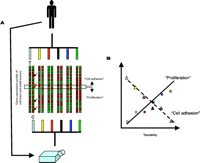
Cell lines and solid tumors may have different patterns of chemosensitivity and gene expression. A. Solid tumors are adherent and inert as compared to their derivatives kept in tissue culture flasks. Gene expression patterns (red and green spots) of tumor samples (T) and cell lines (C) indicate that tumor specimens obtained from biopsies, as well as cancer cell lines maintained in tissue flasks, retain signatures associated with their tissue of origin (illustrated here by schematic dendrograms that cluster tumors according to the tissue of origin, indicated by various colors). A tissue-by-tissue comparison of gene expression patterns between cell lines and solid tumors identifies differently expressed genes in each tumor type [arrows point to genes differently expressed in the first pair-wise comparison with samples obtained from tumor specimens (T) or cell lines (C)]. The expression of a set of genes (highlighted with an open horizontal rectangle) shows consistent differences throughout the tissue types (red arrows point to such genes in the first set of samples). These genes, whose expression is consistently altered as a result of the transition from in vivo to in vitro environments, belong to two major pathways (“Proliferation” and “Cell Adhesion”). Genes belonging to the “Proliferation” pathway are consistently overexpressed in cell lines, whereas the “Cell Adhesion” pathway is downregulated as a result of the adaptation of the cells to the in vitro environment. B. Genes that are differentially expressed in solid tumors and their derivative cell lines show correlation to the tractability of the tumor types. Tissues are known to give rise to cancers with variable response to therapy (axis of tractability). Genes of the “Proliferation” pathway are increasingly overexpressed in more tractable tumor types (expression levels are shown by colored squares, the correlation is indicated by a solid line). Genes of the “Cell Adhesion” pathway are increasingly overexpressed in less tractable tumor types (expression levels are shown by colored triangles, the correlation is indicated by a dashed line). Various colors represent tumor types.
- © American Society for Pharmacology and Experimental Theraputics 2004
References

Michael M. Gottesman, MD, is the Deputy Director for Intramural Research at the NIH and Chief of the Laboratory of Cell Biology, National Cancer Institute, NIH. He attended Harvard College and Harvard Medical School. He and his colleagues identified the human MDR1 gene, which is responsible for the resistance of cancer cells to many of the most common anti-cancer drugs. They have shown that this gene encodes a protein that acts to pump anti-cancer drugs out of drug-resistant human cancers. Dr. Gottesman is a fellow of the AAAS, received the ASPET Award in 1997, and was elected to the Institute of Medicine in 2003. E-mail mgottesman{at}nih.gov; fax 301-402-0450.

Gergely Szakács obtained his MD and PhD degrees at the Semmelweis University in Budapest, Hungary. Dr. Szakács is currently a Visiting Fellow at the National Institutes of Health, USA. E-mail szakacsg{at}mail.nih.gov