
- Institution: Stanford Univ Med Ctr Lane Med Lib/Periodical Dept/Rm L109
- Sign In as Member / Individual
Transcriptional Mechanisms Underlying Addiction-Related Structural Plasticity
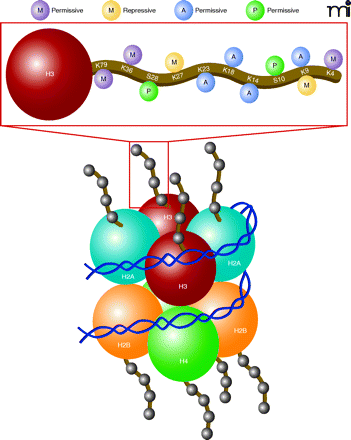
Chromatin modifications on histones regulate gene expression patterns. Chromatin functions to ensure the proper storage, organization and output of genetic information. Shown is the nucleosome core particle, which represents the fundamental repeating transcriptional unit of chromatin, composed of 147 base pairs of DNA wrapped around a core octamer of histone proteins (two copies each of H2A, H2B, H3, and H4, or variants of these proteins). Nucleosomal structural variations are regulated by covalent post-translational modifications of histones, leading to alterations in chromatin compaction leading to more “open” (euchromatic) versus “closed” (heterochromatic) transcriptional states. Combinations of acetylation and numerous other post-translational modifications occurring on N-terminal histone tails (including phosphorylation, methylation, sumoylation, etc.) have been shown to alter chromatin condensation leading to altered levels of gene expression in cells. As shown above, histone modifications that weaken the interaction between histones and DNA, such as histone acetylation at K23, K18, K14, and K9, methylation at K79, K36, and K4, or phosphorylation at S28 and S10, are correlated with transcriptional activation. Likewise, histone deacetylation or histone methylation, which strengthens the electrostatic interaction between histones and DNA, of K27 or K9, promotes a state of transcriptional repression.


