|
 
 |
|
ORIGINAL ARTICLE |
|
|
|
| Year : 2011 | Volume
: 17
| Issue : 3 | Page : 194-200 |
| |
APOA1 gene polymorphisms in the South Asian immigrant population in the United States
Rebecca S Henkhaus1, Sunita Dodani2, Ann M Manzardo1, Merlin G Butler1
1 University of Kansas Medical Center, Departments of Psychiatry and Behavioral Sciences and Pediatrics, 3901 Rainbow Blvd., Kansas City, Kansas, 66160, USA
2 Department of Internal Medicine, 3901 Rainbow Blvd., Kansas City, Kansas, 66160, USA
| Date of Web Publication | 20-Jan-2012 |
Correspondence Address:
Merlin G Butler
3901 Rainbow Blvd, MS 4015, Kansas City, KS, 66160
USA
 Source of Support: This work was partially funded by NIH grants: NHLBI L091476-0, NICHD HD061222, and NICHD HD02528., Conflict of Interest: None  | 2 |
DOI: 10.4103/0971-6866.92103

 Abstract Abstract | | |
Background : Coronary artery disease (CAD) is a leading cause of death in the United States. South Asian immigrants (SAIs) from the Indian subcontinent living in the US are disproportionately at higher risk of CAD than other immigrant populations. Unique genetic factors may predispose SAIs to increased risk of developing CAD when adopting a Western lifestyle including a higher-fat diet, more sedentary behavior and additional gene-environment interactions. SAIs are known to have low levels of the protective high density lipoprotein (HDL) and an altered function for Apo-lipoprotein A-1 (ApoA1), the main protein component of HDL cholesterol. One gene that may be genetically distinctive in this population is APOA1 which codes for ApoA-1 protein, a potentially important contributing factor in the development of CAD.
Materials and Methods : DNA sequencing was performed to determine the status of the seven single-nucleotide polymorphisms (SNPs) in the APOA1 gene from 94 unrelated SAI adults. Genotypes, allelic frequencies, and intragenic linkage disequilibrium of the APOA1 SNPs were calculated.
Results : Several polymorphisms and patterns were common among persons of south Asian ethnicity. Frequencies for SNPs T655C, T756C and T1001C were found to be different than those reported in European Caucasian individuals. Linkage disequilibrium was found to be present between most (13 of 15) SNP pairings indicating common inheritance patterns.
Conclusions : SAIs showed variability in the sequence of the APOA1 gene and linkage disequilibrium for most SNPS. This pattern of APOA1 SNPs may contribute to decreased levels of HDL cholesterol reported in SAIs, leading to an increased risk for developing CAD in this population.
Keywords: APOA1 , coronary artery disease, linkage disequilibrium, single nucleotide gene polymorphisms, south Asian immigrants.
How to cite this article:
Henkhaus RS, Dodani S, Manzardo AM, Butler MG. APOA1 gene polymorphisms in the South Asian immigrant population in the United States. Indian J Hum Genet 2011;17:194-200 |
How to cite this URL:
Henkhaus RS, Dodani S, Manzardo AM, Butler MG. APOA1 gene polymorphisms in the South Asian immigrant population in the United States. Indian J Hum Genet [serial online] 2011 [cited 2016 May 13];17:194-200. Available from: http://www.ijhg.com/text.asp?2011/17/3/194/92103 |
 Introduction Introduction | |  |
Genetic haplotypes and single nucleotide polymorphisms (SNPs) in the human genomic DNA sequence frequently differ among ethnic groups. [1],[2] It is imperative to study and document genetic variation among ethnic groups, especially as diagnosis and treatment of disease moves increasingly toward more personalized, specific approaches to medical care. Studying the genetics of geographically diverse groups of people has revealed important differences at the molecular level. A classic example is a single base pair change in the hemoglobin gene which causes sickle cell disease and occurs more often in persons of direct African descent. [3] Many of these genetic discoveries have played critical roles in the diagnosis and treatment of a variety of conditions from classically inherited diseases to more complex disorders such as cancer and heart disease. For example, South Asian immigrants (SAIs- people with ancestors from the Indian subcontinent, i.e. India, Pakistan, Bangladesh, Nepal, Bhutan, and Sri Lanka) living abroad in regions with more Westernized cultures such as the United Kingdom, Canada and the United States have experienced higher rates of heart disease than other ethnic groups. [4],[5] Additionally, in the SAI population, the highest rates of mortality occur from coronary artery disease (CAD) and at a younger age than in other ethnic groups. [6],[7],[8],[9] SAIs constitute the second largest Asian immigrant population as well as the fastest growing ethnic immigrant population in the United States.
Many factors contribute to CAD. Clearly, an individual's environment and behaviors contribute, but genetic factors play a large role especially in the immigrant population. Factors that may be partially influenced by an individual's behavior, such as BMI, cholesterol levels and blood pressure are also largely determined by genetics. [10] However, the factor that seems to be of major concern among the population of SAIs is the triglyceride level and high-density lipoprotein (HDL) to low-density lipoprotein (LDL) ratios. When compared to similarly age-matched Caucasians in the United States, South Asian men have lower levels of hypertension, tobacco usage, and obesity. [11] But, the same study demonstrated that SAIs in the United States have dyslipidemia (high LDL and triglyceride levels, accompanied by low HDL levels) and a greater prevalence of noninsulin-dependent diabetes mellitus, or type 2 diabetes. [11] High levels of HDL cholesterol and a high ratio of HDL to LDL cholesterol are protective against heart disease. Low HDL levels alone or accompanied by high triglyceride and high lipoprotein levels have been documented in the SAIs Indian population. [12] The size of HDL particles has been shown to be smaller in South Asian men; larger HDL particles may be more effective at scavenging cellular cholesterol and facilitating its removal from circulation. Smaller HDL particles are not as useful in facilitating reverse uptake of circulating cholesterol. [13],[14] As cholesterol and specifically low HDL cholesterol levels appear to play a central role in the development of heart disease in South Asian immigrants, this article explores the underlying genetics of this phenomenon and the role of APOA1 gene polymorphisms in our SAI study population.
The APOA1 gene encodes the protein, ApoA-1, which is the principle protein component of HDL cholesterol. Low serum levels of ApoA-1 correlate with the occurrence of heart disease. [15] The APOA1 gene is contained within the APOA1/C3/A4/A5 cluster on chromosome 11q23, with more than 182 distinct SNPs identified in this gene cluster region. [16] Analysis of the APOA1/C3/A4/A5 gene cluster has shown linkage disequilibrium within a 150 kb span of the region. [17] Previously SAIs have been reported to carry six specific SNPs in the APOA1 gene [Table 1] located in the intronic regions of the APOA1 gene. [18] Although these SNPs are not located in the coding regions of the gene, they potentially can impact APOA1 gene expression. Additionally, we analyzed a separate SNP in the 5' UTR (-75G>A) region that is present in an estimated 11-35% of the population, in a variety of racial and ethnic groups, and shown to decrease APOA1 gene expression and ApoA-1 protein translation. [16],[19],[20]
The characterization of the APOA1 gene is of importance due to the role of HDL cholesterol and ApoA-1 levels in the development of heart disease in the SAI population. Thus, we determined the frequency and pattern of occurrence of SNPs in the APOA1 gene in the SAI population. Certain polymorphic alleles were found to be more common than the so called wild-type alleles, using the National Center for Biotechnology Information (NCBI) database APOA1 sequence (NC_000011.9), highlighting the fact that the consensus genomic DNA reference sequences do not always reflect every ethnic population studied.
 Materials and Methods Materials and Methods | |  |
Sample collection and demographics: Adult SAI males and females (n=74, age range of 35-65 years) representing first and second generations living in the United States were recruited in the Kansas City Metro area at local places of worship following the distribution of flyers and announcements outlining the purpose, rationale, and design of the study. Informed consents were obtained from each subject. This study was approved by the University of Kansas Medical Center Human Subjects Committee. Adult SAI adult males and females (n=20) were recruited and approved by the Medical College of Georgia. [18]
Blood collection and DNA extraction: Whole blood was obtained in 10 ml citrate anti-coagulated vacutainer blood tubes by trained phlebotomists. Genomic DNA was isolated routinely from the whole blood using the FlexiGene DNA Kit (Qiagen, Valencia, CA).
DNA sequencing of APOA1 gene: PCR amplification and sequencing of the APOA1 gene encompassing the SNPs to be analyzed was performed as previously described. [21] Briefly, a 1683bp fragment of the APOA1 gene was amplified using the following primers: forward 5' CACAATGGACAATGGCAACT 3' and reverse 5' CCAGATCCTTGCTCATCTCC 3'. The fragment was sequenced using a series of two sequencing primers (first sequencing primer: 5' CTTGACCCCTGCCCTGCAGC 3'; second sequencing primer: 5' CGGCAGAGACTATGTGTCCCAG 3') to encompass all seven SNPs. Sequencing data for each subject were provided in the form of raw electropherogram files from which DNA sequences were derived using Ridom Trace Edit software (Ridom Bioinformatics, Würzburg, Germany). All SNPs were determined and verified both from the derived DNA sequence and visual inspection of the electropherogram. Results were analyzed by comparative NCBI BLAST sequence analysis. The reference sequence used was NCBI RefSeq NC_000011.9, derived from the Genome Reference Consortium Human Build 37 (GRCh37), Primary Assembly ( http://www.ncbi.nlm.nih.gov/gene ).
Statistical analysis: Intragenic linkage disequilibrium (LD) was determined by comparing haplotypes frequencies using standard LD calculations. [22] The number of subjects with each allele pairing haplotype, represented as A 1 B 1 (wild-type/wild-type), A 1 B 2 (wild-type/SNP), A 2 B 1 (SNP/wild-type), and A 2 B 2 (SNP/SNP) were counted, then haplotype frequencies were determined by dividing each haplotypes count by the total number of alleles: x 11 = A 1 B 1 /2n; x 12 = A 1 B 2 /2n; x 21 = A 2 B 1 /2n; x 22 = A 2 B 2 /2n. Allele frequencies were then calculated from the haplotype frequencies: p 1 = x 11 +x 12 ; p 2 = x 21 +x 22 ; q 1 = x 11 +x 21 ; q 2 = x 12 +x 22 . Linkage disequilibrium (D) was then calculated: D = x 11 -(p 1 q 1 ). Using the linkage disequilibrium values, Chi-squared calculations were performed: X 2 = 2nD 2 /(p 1 p 2 q 1 q 2 ). Significance was determined using a non-directional 2-tailed Chi-square distribution with one degree of freedom. Values greater than 6.64 have a significance of P < 0.01 and values greater than 10.83 have a significance of P < 0.001.
 Results Results | |  |
Genotype analysis and determination of allele frequency: The allele and genotype frequencies for each SNP are listed in [Table 2] and were recently reported in relation to metabolic syndrome. [21] SNPs T655C, T756C and T1001C were found to be the most common, with polymorphic allelic frequencies of 70.2, 70.7 and 67.0%, respectively. These three SNPs also nearly always occurred together in the same subject. For example, all subjects who carried at least one polymorphic allele at position 655 also carried a polymorphism at positions 756 and 1001, while only one subject carried the polymorphism at position 1001, but not at positions 655 or 756. Although the C938T and C1149T SNPs were contained within the same APOA1 gene intron (between exons 3 and 4), polymorphic alleles for these SNPs did not necessarily coincide with SNPs at positions 655, 756 and 1001. The C938T and C1149T SNPs were also far less common, occurring in only 11.2 and 4.3% of subjects, respectively, with no subjects exhibiting homozygosity for these polymorphisms. The C319T SNP was found in 27.7% of subjects. No apparent association was detected with the occurrence of any other SNP. The SNP (G-75A) located in the 5'UTR, near the promoter region was found in 20.3% of 74 SAI subjects studied for the SNP.
Patterns of SNP occurrence in SAI subjects: Patterns of SNPs were analyzed to discover the most common concurrent groupings of SNPs [Figure 1]a. Slightly more than one half of the subjects (51.2% or 43 of 82) who carried the 655/756 polymorphic alleles also carried the T319C polymorphism. Three patterns of SNP occurrence covered 84.1% (69 of 82) of the subjects studied having more than one polymorphic allele. These patterns included polymorphisms at positions 655/756/1001, 319/655/756/1001 or 655/756/938/1001. The remaining 13 subjects with more than one SNP comprised all other patterns making them far less common. Overall, there were nine separate patterns of polymorphism occurrence in the subjects studied [Figure 1]a. Of the nine patterns, each with six SNPs, 21 of 54 most prevalent alleles were wild-type and 33 were polymorphic, making the polymorphisms more common than wild-type alleles in the SAI population.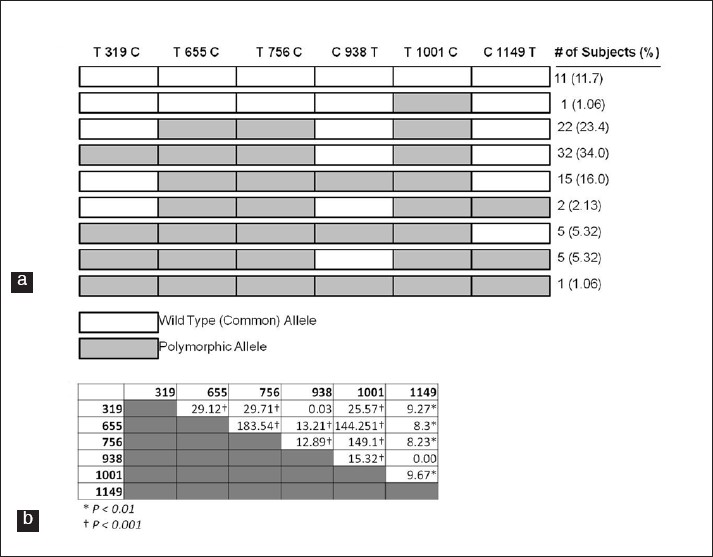 | Figure 1: Linkage of APOA1 Polymorphisms: (a) Concurrence of SNPs in the SAI study population (n=94). Nucleotide loci refer to the APOA1 NCBI reference sequence (NC_000011.9). (b) Chi-squared values calculated from linkage disequilibrium data for indicated SNP pairings (calculations described in Materials and Methods section). Significant values indicate SNPs in linkage disequilibrium (*P < 0.01; **P < 0.001).
Click here to view |
Linkage disequilibrium and Hardy-Weinberg analysis: Linkage disequilibrium (LD) analysis was performed on each combination of the six SNPs within the APOA1 gene [Figure 1]b. All alleles were found to be significantly in LD except for two pairings: 319 vs. 938 and 938 vs. 1149. Three SNPs (655, 756 and 1001) were found to have a very high degree of LD as compared to the other pairings. These data indicate that SNP 938 is inherited in a more independent manner than the other SNPs within the APOA1 gene while SNPs 655, 756 and 1001 had the highest degree of LD. Given the observed frequency of each allele, none were significantly different from expected Hardy-Weinberg equilibrium (data not shown).
 Discussion Discussion | |  |
Gene sequencing and SNP assays are becoming easier and more widely available, leading to establishment of personalized medicine approaches in treating diseases. Genetic differences between ethnic groups will most certainly reveal the causes for differences in susceptibility to a variety of diseases. Characterization of polymorphisms in the APOA1 gene may well shed light on why South Asians are disproportionately affected by heart disease with ApoA-1 protein levels being the single most powerful predictor for the development of heart disease. [23],[24],[25] This study further shows that the sequence of the APOA1 gene in SAIs is commonly divergent from the wild-type gene sequence, the NCBI reference sequence NC_000011.9, established from the Human Genome Reference Consortium (GRCh37) primary assembly. This most recent sequence assembly was compiled using data collected from multiple individuals, likely from a different (non-South Asian) ethnic background. It should be noted that the Celera Assembly reference for APOA1 indicates different wild-type alleles for the SNPs analyzed in this study: G-75A, C319T, C655T, C756T, C938T, C1001T and C1149T. This difference in reference assembly sequences confirms that these polymorphisms are highly diverse among individuals, underscoring the need to elucidate and characterize the sequence differences among ethnic groups and determine the impact of SNPs on ApoA-1 protein production.
The SNPs analyzed in our study have been examined in a variety of ethnic groups. For example, the SNP located in the 5'UTR (G-75A) has been the most widely studied. A meta-analysis was previously performed for the minor-allele frequency (MAF) determination with G-75A polymorphism across a range of ethnic groups. [19] For our discussion, the term MAF refers to the frequency of the polymorphic allele, although it may not represent the allele with the lower frequency. Caucasians primarily of northern and western European descent have an MAF of 0.11-0.21. [19] Individuals of Asian (Japan and Singapore) descent have an MAF ranging from 0.14 to 0.28. [19] Our study demonstrated that SAIs have an MAF of 0.20 for this SNP.
The other six SNPs within the APOA1 gene have also been studied in multiple ethnic groups. Allele frequencies obtained from the SNP database (dbSNP) ( http://www.ncbi.nlm.nih.gov/snp ) [26] for studies representing diverse ethnic groups from Europe, Africa, Thailand, China and India have been summarized in [Table 3]. Notable differences from our study include the higher prevalence of polymorphic allele in persons of European heritage at position 319, and the decreased frequency of the polymorphic allele in Thai individuals at positions 655, 756 and 1001. The alleles analyzed in individuals of Indian ethnicity living in the United States were all highly similar to the SNP allele frequencies observed in our SAI population [Table 3].
In addition to determining the SNP frequencies for the SAI population, the intragenic linkage disequilibrium was also analyzed using established protocols for the six SNPs within the APOA1 gene. Of the 15 pairwise analyses, 13 were found to be in linkage disequilibrium. The SNPs at locations 655, 756 and 1001 had the highest degree of linkage while the SNPs at locations 319, 1149, and especially 938 had a more independent inheritance pattern [Figure 1]. A study by Qi et al, [27] demonstrated that common intragenic haplotypes can be predictive of clinical outcomes. Their study reported a haplotype in the human perilipin gene (PLIN) common to Malays and Indian populations that was significantly correlated with increased obesity risk as compared to individuals of Chinese descent, with all three ethnic populations living in Singapore. [27] A similar study could eventually be carried out with SAI subjects compared with differing ethnic populations, assessing the correlation between APOA1 haplotypes and relative risk for developing CAD.
These data provide an excellent basis for further studies of APOA1 gene expression in the SAI population. Although the G-75A SNP has previously been studied in relation to gene expression, the other SNPs have not. Research has shown that the presence of the polymorphic allele at the G-75A SNP is associated with lower APOA1 gene expression. [20] A study conducted on individuals from Northern India showed that significantly lower levels of both plasma ApoA-1 and HDL cholesterol were present in persons carrying the A allele, as opposed to those with the GG (wild-type) genotype. [28] The other SNPs may have a comparable impact on APOA1 gene expression, and further explorations are currently underway using our study population. The initial study which reported the presence of the six intronic SNPs in the SAI population also demonstrated that the C938T SNP is significantly correlated with low plasma HDL cholesterol (<40 mg) levels. [18] Furthermore, a study performed comparing first generation American-born adults of South Asian heritage with American-born adults of European heritage demonstrated that the South Asian individuals had lower HDL levels, but higher LDL and triglyceride levels; yet the South Asians had lower BMI. [29]
A recently published study on this cohort of SAI subjects revealed a statistically significant correlation between three of the APOA1 SNPs (C655T, C756T, and C1001T) and metabolic syndrome (MS) with positive odds ratios ranging from 2.5 to 3.0 (CI>95%). [21] Negative odds ratios (CI>95%) were also observed for HDL cholesterol, Apo-A1 protein and lipoprotein(a), indicating that high levels of these proteins, all of which are influenced by APOA1 gene expression patterns and protective against MS. [21] These findings may be of even greater relevance given the data reported herein that the three significantly correlated SNPs have the highest degree of LD, indicating a conserved hereditary pattern. The occurrence of MS was also significantly correlated with the presence of pro-inflammatory HDL (dys-HDL) as well as increased common carotid artery intima-media thickness (CCA-IMT), a surrogate marker for CAD. [21] Given that SAIs are known to carry a disproportionately high risk for CAD, there is need to explore and understand gene-environment interactions. [6],[7],[8],[9] This increased risk seems to be primarily due to dyslipidemia, as opposed to other common risk factors such as obesity and high blood pressure. Apo-A1 is a key component of HDL and genetic changes that affect APOA1 expression may well play an important role in CAD as well as MS.
Clearly, genetic factors play a large role in determining cholesterol levels and ratios in all human populations. Family and twin studies have shown that genetic heritability accounts for nearly two-thirds of the variability observed between individuals' HDL and ApoA-1 levels. [28],[30] In the SAI population, these genetic factors may lead to an increased incidence and mortality resulting from heart disease. While living in the countries and cultures of South Asia, the propensity toward dyslipidemia may not lead to a selective disadvantage. The diet and level of activity more common in individuals living in southern Asia may not interplay with the genetic predisposition to lead to common occurrence of heart disease, as opposed to the more Western lifestyle of the United States, Canada and the United Kingdom with greater access to animal products, leading to more consumption of cholesterol in the diet. This research and future related studies should lend crucial insight into the causes of heart disease in South Asians living in Westernized societies, and certainly will provide the knowledge needed for more targeted and effective prevention and treatment options.
 Acknowledgements Acknowledgements | |  |
We would like to thank the participants who contributed their valuable time to further our knowledge. We are also thankful to the Hindu temples in the Kansas City Metro area for supporting our study and helped with subject recruitment. We thank the study coordinator Ms. Kuntal Shastri for administrative assistance, as well as Yan Bin Dong and Haidong Zhu at the Medical College of Georgia for their assistance in the preparation and sequencing of previous SAI DNA samples.
 References References | |  |
| 1. | Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung HC, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature 2008;451:998-1003. 
[PUBMED] [FULLTEXT] |
| 2. | Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science 2008;319:1100-4. 
[PUBMED] [FULLTEXT] |
| 3. | Crepeau RH, Edelstein SJ, Szalay M, Benesch RE, Benesch R, Kwong S, et al. Sickle cell hemoglobin fiber structure altered by alpha-chain mutation. Proc Natl Acad Sci U S A 1981;78:1406-10. 
[PUBMED] [FULLTEXT] |
| 4. | Enas EA, Senthilkumar A. Coronary artery disease in Asian Indians: An update and review. Internet J Cardiol 2002. p. 1. 
|
| 5. | McKeigue PM, Miller GJ, Marmot MG. Coronary heart disease in south Asians overseas: A review. J Clin Epidemiol 1989;42:597-609. 
[PUBMED] [FULLTEXT] |
| 6. | Gupta M, Singh N, Verma S. South Asians and cardiovascular risk: What clinicians should know. Circulation 2006;113:e924-9. 
[PUBMED] [FULLTEXT] |
| 7. | Klatsky AL, Tekawa I, Armstrong MA, Sidney S. The risk of hospitalization for ischemic heart disease among Asian Americans in northern California. Am J Public Health 1994;84:1672-5. 
[PUBMED] [FULLTEXT] |
| 8. | Sheth T, Nair C, Nargundkar M, Anand S, Yusuf S. Cardiovascular and cancer mortality among Canadians of European, south Asian and Chinese origin from 1979 to 1993: an analysis of 1.2 million deaths. CMAJ 1999;161:132-8. 
[PUBMED] [FULLTEXT] |
| 9. | Singh N, Gupta M. Clinical characteristics of South Asian patients hospitalized with heart failure. Ethn Dis Autumn 2005;15:615-9. 
|
| 10. | Neufeld HN, Goldbourt U. Coronary heart disease: Genetic aspects. Circulation 1983;67:943-4. 
[PUBMED] [FULLTEXT] |
| 11. | Enas EA, Garg A, Davidson MA, Nair VM, Huet BA, Yusuf S. Coronary heart disease and its risk factors in first-generation immigrant Asian Indians to the United States of America. Indian Heart J 1996;48:343-53. 
[PUBMED] |
| 12. | Dhawan J. Coronary heart disease risks in Asian Indians. Curr Opin Lipidol 1996;7:196-8. 
[PUBMED] |
| 13. | Bhalodkar NC, Blum S, Rana T, Bhalodkar A, Kitchappa R, Kim KS, et al. Comparison of levels of large and small high-density lipoprotein cholesterol in Asian Indian men compared with Caucasian men in the Framingham Offspring Study. Am J Cardiol 2004;94:1561-3. 
[PUBMED] [FULLTEXT] |
| 14. | Johansson J, Carlson LA, Landou C, Hamsten A. High density lipoproteins and coronary atherosclerosis: A strong inverse relation with the largest particles is confined to normotriglyceridemic patients. Arterioscler Thromb 1991;11:174-82. 
[PUBMED] [FULLTEXT] |
| 15. | Brunzell JD, Sniderman AD, Albers JJ, Kwiterovich PO Jr. Apoproteins B and A-I and coronary artery disease in humans. Arteriosclerosis 1984;4:79-83. 
[PUBMED] [FULLTEXT] |
| 16. | Shanker J, Perumal G, Rao VS, Khadrinarasimhiah NB, John S, Hebbagodi S, et al. Genetic studies on the APOA1-C3-A5 gene cluster in Asian Indians with premature coronary artery disease. Lipids Health Dis 2008;7:33. 
[PUBMED] [FULLTEXT] |
| 17. | Olivier M, Wang X, Cole R, Gau B, Kim J, Rubin EM, et al. Haplotype analysis of the apolipoprotein gene cluster on human chromosome 11. Genomics 2004;83:912-23. 
[PUBMED] [FULLTEXT] |
| 18. | Dodani S, Dong Y, Zhu H, George V. Can novel Apo A-1 polymorphisms be responsible for low HDL in South Asian immigrants? Indian J Hum Genet 2008;14:9-15. 
[PUBMED]  |
| 19. | Juo SH, Wyszynski DF, Beaty TH, Huang HY, Bailey-Wilson JE. Mild association between the A/G polymorphism in the promoter of the apolipoprotein A-I gene and apolipoprotein A-I levels: A meta-analysis. Am J Med Genet 1999;82:235-41. 
[PUBMED] [FULLTEXT] |
| 20. | Smith JD, Brinton EA, Breslow JL. Polymorphism in the human apolipoprotein A-I gene promoter region: Association of the minor allele with decreased production rate in vivo and promoter activity in vitro. J Clin Invest 1992;89:1796-800. 
[PUBMED] [FULLTEXT] |
| 21. | Dodani S, Henkhaus R, Wick J, Vacek J, Gupta K, Dong L, et al. Metabolic syndrome in South Asian immigrants: More than low HDL requiring aggressive management. Lipids Health Dis 2011;10:45. 
|
| 22. | Devlin B, Risch N. A comparison of linkage disequilibrium measures for fine-scale mapping. Genomics 1995;29:311-22. 
[PUBMED] [FULLTEXT] |
| 23. | Andrikoula M, McDowell IF. The contribution of ApoB and APOA1 measurements to cardiovascular risk assessment. Diabetes Obes Metab 2008;10:271-8. 
[PUBMED] [FULLTEXT] |
| 24. | Garfagnini A, Devoto G, Rosselli P, Boggiano P, Venturini M. Relationship between HDL-cholesterol and apolipoprotein A1 and the severity of coronary artery disease. Eur Heart J 1995;16:465-70. 
[PUBMED] [FULLTEXT] |
| 25. | O'Brien T, Nguyen TT, Hallaway BJ, Hodge D, Bailey K, Holmes D, et al. The role of lipoprotein A-I and lipoprotein A-I/A-II in predicting coronary artery disease. Arterioscler Thromb Vasc Biol 1995;15:228-31. 
[PUBMED] [FULLTEXT] |
| 26. | Entrez SNP Database. Available from: http://www.ncbi.nlm.nih.gov/snp. [accessed on 2010 Apr 1]. 
|
| 27. | Qi L, Tai ES, Tan CE, Shen H, Chew SK, Greenberg AS, et al. Intragenic linkage disequilibrium structure of the human perilipin gene (PLIN) and haplotype association with increased obesity risk in a multiethnic Asian population. J Mol Med 2005;83:448-56. 
[PUBMED] [FULLTEXT] |
| 28. | Chhabra S, Narang R, Lakshmy R, Das N. APOA1-75 G to A substitution associated with severe forms of CAD, lower levels of HDL and apoA-I among northern Indians. Dis Markers 2005;21:169-74. 
[PUBMED] [FULLTEXT] |
| 29. | Kalhan R, Puthawala K, Agarwal S, Amini SB, Kalhan SC. Altered lipid profile, leptin, insulin, and anthropometry in offspring of South Asian immigrants in the United States. Metabolism 2001;50:1197-202. 
[PUBMED] [FULLTEXT] |
| 30. | Dodani S, Kaur R, Reddy S, Reed GL, Navab M, George V. Can dysfunctional HDL explain high coronary artery disease risk in South Asians? Int J Cardiol 2008;129:125-32. 
[PUBMED] [FULLTEXT] |
[Figure 1]
[Table 1], [Table 2], [Table 3]
| This article has been cited by | | 1 |
Re-sequencing of the APOAI promoter region and the genetic association of the -75G?>?A polymorphism with increased cholesterol and low density lipoprotein levels among a sample of the Kuwaiti population |
|
| Suzanne A Al-Bustan,Ahmad E Al-Serri,Babitha G Annice,Majed A Alnaqeeb,Ghada A Ebrahim | | BMC Medical Genetics. 2013; 14(1): 90 | | [Pubmed] | [DOI] | |
|
 |
|






