|
 
 |
|
ORIGINAL ARTICLE |
|
|
|
| Year : 2013 | Volume
: 19
| Issue : 1 | Page : 54-57 |
| |
Investigation of the A1555G mutation in mitochondrial DNA (MT-RNR1) in groups of Brazilian individuals with nonsyndromic deafness and normal-hearing
Karina Bezerra Salomão, Christiane Maria Ayo, Valter Augusto Della-Rosa
Department of Biotechnology, Genetics and Cellular Biology, State University of Maringa, Paraná, Brazil
| Date of Web Publication | 4-Jun-2013 |
Correspondence Address:
Valter Augusto Della-Rosa
Department of Biotechnology, Genetics and Cellular Biology, State University of Maringa, Colombo Avenue, 5790, Maringa-PR, 87020-900
Brazil
 Source of Support: This study was supported by Brazilian Agencies CNPq (Conselho Nacional de Desenvolvimento Tecnológico) and FA (Fundação Araucária do Estado do Paraná),, Conflict of Interest: None
DOI: 10.4103/0971-6866.112888

 Abstract Abstract | | |
Background: Mutations of mitochondrial DNA were described into two genes: The mitochondrially encoded 12S RNA (MT-RNR1) and the mitochondrially encoded tRNA serine ucn (MT-TS1). The A1555G mutation in MT-RNR1 gene is a frequent cause of deafness in different countries.
Aim: The aim of this work was to investigate the frequency of the A1555G mutation in the MT-RNR1 gene in the mitochondrial DNA in Brazilians individuals with nonsyndromic deafness, and listeners.
Materials and Methods: DNA samples were submitted to polymerase chain reaction and to posterior digestion with the Hae III enzyme.
Results: Seventy eight (78) DNA samples of deaf individuals were analyzed; 75 showed normality in the region investigated, two samples (2.5%) showed the T1291C substitution, which is not related to the cause of deafness, and one sample (1.3%) showed the A1555G mutation. Among the 70 non-impaired individuals no A1555G mutation or T1291C substitution was found.
Conclusion: We can affirm that A1555G mutation is not prevalent, or it must be very rare in normal-hearing subjects in the State of Paranα, the south region of Brazil. The A1555G mutation frequency (1.3%) found in individual with nonsyndromic deafness is similar to those found in other populations, with nonsyndromic deafness. Consequently, it should be examined in deafness diagnosis. The investigation of the A1555G mutation can contribute towards the determination of the nonsyndromic deafness etiology, hence, contributing to the correct genetic counseling process.
Keywords: A1555G mutation, deafness, hearing loss, listeners, mitochondrial DNA
How to cite this article:
Salomão KB, Ayo CM, Della-Rosa VA. Investigation of the A1555G mutation in mitochondrial DNA (MT-RNR1) in groups of Brazilian individuals with nonsyndromic deafness and normal-hearing. Indian J Hum Genet 2013;19:54-7 |
How to cite this URL:
Salomão KB, Ayo CM, Della-Rosa VA. Investigation of the A1555G mutation in mitochondrial DNA (MT-RNR1) in groups of Brazilian individuals with nonsyndromic deafness and normal-hearing. Indian J Hum Genet [serial online] 2013 [cited 2016 May 24];19:54-7. Available from: http://www.ijhg.com/text.asp?2013/19/1/54/112888 |
 Introduction Introduction | |  |
Hearing loss is commonly divided into nonsyndromic and syndromic. Nonsyndromic deafness is that without additional phenotypes, and syndromic is a hearing loss which is part of a group of signals, constituting a syndrome. Multiple loci which cause deafness have been mapped and described in autosomal dominant nonsyndromic deafness loci (DFNA), autosomal recessive nonsyndromic deafness loci (DFNB) and X linked nonsyndromic deafness (DFN).[1] Mutations of mitochondrial DNA (mtDNA) were described into two genes: Mitocondrially encoded 12S RNA (MT-RNR1) and the mitocondrially encoded tRNA serine ucn (MT-TS1) associated with nonsyndromic deafness. The A1555G mutation in the MT-RNR1 gene is the most frequently described cause of deafness. [2]
Mitochondrial mutations can be associated to both nonsyndromic and syndromic hearing loss, showing heteroplasmya in the last one, certainly, due to the lethality affecting other organs. [3]
The A1555G mutation in the MT-RNR1 (12S rRNA) causes individuals' susceptibility to deafness induced by aminoglycosides. It was estimated that in the USA this mutation was responsible for approximately 15% of all cases of deafness induced by aminoglycosides. [4]
In the A1555G mutation by the substitution of nitrogenated bases, a new pair of C-G bases appear in human the 12S rRNA gene, making it similar to the region that corresponds to the 16S rRNA gene of Escherichia More Details coli, bacterial decoding region of rRNA, in which the aminoglycoside link results in protein translation errors, and subsequently in bacterial death. Mutations in the sub-unit 12S of rRNA probably alter the secondary structure of the molecules making it very similar to the 16S rRNA molecules of the E. coli. As the 16S rRNA molecule is the main target of the aminoglycoside action, this can explain the aminoglycoside increased effect in individuals who present this mutation, leading to hearing loss. [5]
The phenotypical result of the mitochondrial A1555G mutation shows variation between severe deafness, moderate progressive hearing loss, and normal hearing among relatives who share the same maternal lineage. The incomplete penetration and varied expressivity of hearing loss associated to the A1555G mutation are related to the interaction among genetic factors, modified nuclear genes, mitochondrial haplotype, and environmental factors such as aminoglycosides. [6]
The mitochondrial mutations have great importance in the etiology of the sensorineural hearing deficiency, and for the fact that Brazil is a multiregional and multi-ethnic country; the frequency of this mutation is poorly known. The present study had the aim to investigate the frequency of the A1555G mutation in the MT-RNR1 gene in the mitochondrial DNA, in people with nonsyndromic deafness showing negative results for the 35delG, 167delT and 235delC mutations in the gap junction protein beta 2 gene (GJB2) and in normal hearing subjects from the State of Paranα, in the south region of Brazil.
 Materials and Methods Materials and Methods | |  |
Subjects
The deafness group was constituted of 78 patients with nonsyndromic deafness. Their ages varied from 3 months to 76 years old (age average = 16.4 years), 41 males and 37 females. All of them had already been investigated for the 35delG, 167delT and 235delC mutations in the GJB2 gene, showing negative results.
The non-impaired group was constituted of 70 people, 28 males and 42 females, with ages ranging from 18 to 58 years old (average age = 24.18 years), with no history of hearing loss or deafness.
The research was approved by the Permanent Committee of Ethics in Research with Human Beings, of the State University of Maringα.
Molecular genetics analysis
The mitochondrial A1555G mutation test was adapted from Estivill et al.[7] technique, consisting of polymerase chain reaction (PCR) followed by the digestion of a product with the Hae III (10 U/μl) enzyme of restriction. For the PCR reaction, it was used between 100 ng and 200 ng of total DNA; 0.4 μM of each primer 5' GCT CAG CCT ATA TAC CGC CAT CTT CAG CAA 3' and 5' TTT CCA GTA CAC TTA CCA TGT TAC GAC TGG 3'; 1 U of Taq polymerase (Invitrogen); 1.5 mM MgCl2; 20 mM TRIS pH = 8.4 and 50 mM KCl, 0.2 mM dNTPs (Invitrogen), with final volume of 25 μl, with an initial denaturation at 95°C for 5 min, followed by 39 cycles at 94°C of 30 s; 53°C, 1 min; 72°C, 2 min, and finally the extension at 72°C, 10 min; the phases were executed in the Eppendorf Thermocycler.
The digestion of the PCR product was done with the Hae III enzyme. Electrophoresis was done in polyacrylamide gel at 6%, stained with SYBR® Gold nucleic acid stain (Invitrogen). The samples with no mutation showed two bands: 216 bp and 123 bp; the one with mutation showed three bands: 216 bp, 93 bp and 30 bp, the last one is not usually visible.
 Results Results | |  |
From the 78 analyzed DNA samples of deaf patients, 75 showed normality in the investigated region. In one subject (1.3%) it was found the A1555G mutation, and two subjects presented the T1291C substitution, which shows the 173, 123 and 43 bp fragments. The last one was not visualized [Figure 1]. Among 70 normal-hearing individuals, it was not found the A1555G mutation or the T1291C substitution.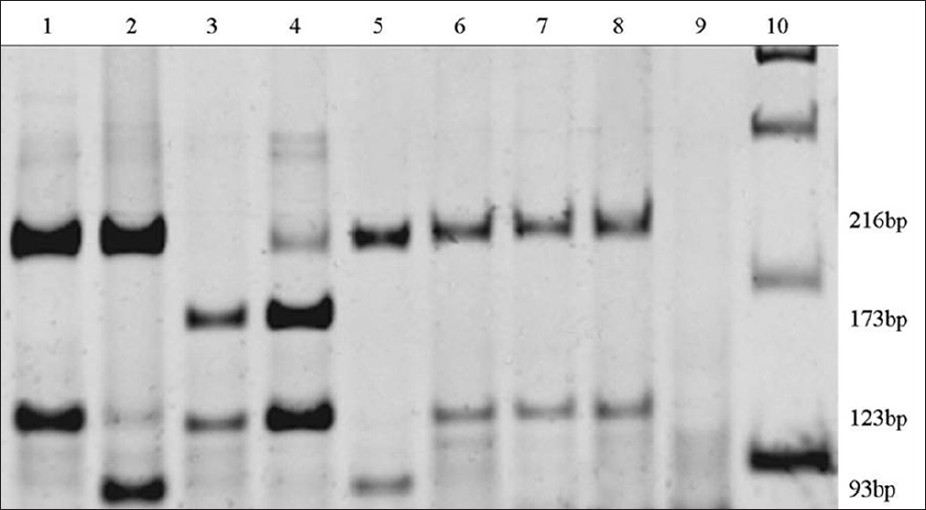 | Figure 1: Polyacrylamide gel at 6%, stained with SYBR® Gold nucleic acid stain: (1) Normal subject control; (2) deaf subject control, with the A1555G mutation; (3 and 4) deaf patients with polymorphism T1291C; (5) deaf patient with the A1555G mutation; (6, 7 and 8) normal hearing patients with no mutation; (9) control without DNA and (10) molecular marker (ladder 100 bp)
Click here to view |
 Discussion Discussion | |  |
The data in the present study contribute to a better knowledge of the A1555G mutation in subjects with nonsyndromic deafness as well as among to the normal hearing population in the State of Paranα, in Brazil. This mutation occurs in different ethnical groups [Table 1] having among the nonsyndromic deafness the frequencies of: 3% in Japanese, [8] 0.7% in German, 2.4% in Polish, [9] 5.3% in Southeast Asian, [10] 2.4% in Danish. [11] However, it is rare or absent in other groups such as Mexicans [12] and Qatari. [13]
Many studies have been carried out in Brazil to attain a better knowledge of the nonsyndromic deafness frequency in the deaf population. In a special care school, in the great Sγo Paulo, Brazil, it was concluded that 3% of the deafness cases were due to mitochondrial mutations. [14] In another research conducted in Sγo Paulo, Brazil, 2% of the individuals with nonsyndromic hearing loss had the mitochondrial A1555G [15] mutation. In our study, the frequency for the A1555G mutation was 1.3% (1 in 78). In another study with 27 deaf participants, in Brazil, the A1555G was not found. [16] The majority of our patients had pre-lingual deafness; a characteristic found in the majority of the Brazilian studies. The clinical presentation of deafness in deafs and their relatives with the A1555G mutation varies according to severity, progression, and initial ages, according to Estivill and collaborators in 1998. [7]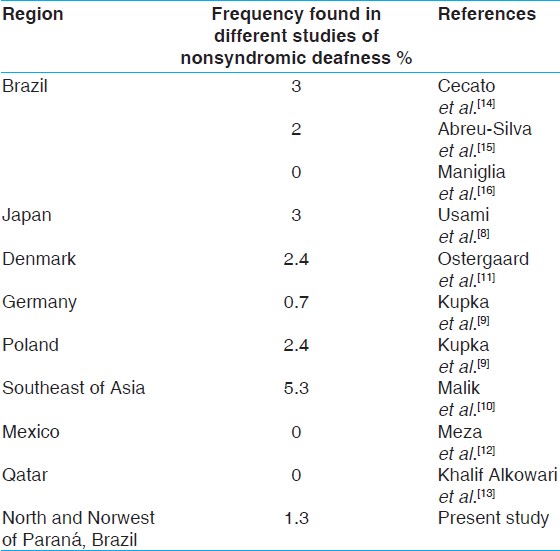 | Table 1: The A1555G mutation frequency in nonsyndromic deafness of studies in different regions
Click here to view |
The affected subject with A1555G found in this study had pre-lingual deafness, and it was diagnosed at the age of 1 year and a half. The aminoglycoside treatment was not reported, although he had an affected aunt on the mother's side.
The T1291C substitution found in two individuals, in the deaf sample of this study, was also reported by different authors as polymorphism with no pathological consequences (MITOMAP database, [2] ). For Abreu-Silva et al.[15] who found it in 2% of the hearing impaired subjects, and 1% in the African decent hearing subjects, the T1291C substitution was not related to the cause of deafness. However, in a study carried out by Ballana et al.,[6] the T1291C change was detected in a Cuban family affected by nonsyndromic sensorineural deafness, perfectly segregating with hearing loss. The researchers hypothesize that the changes in the primary structure of the DNA, as for example the T1243C and the T1291C changes, correspond to the alterations in the 12S rRNA secondary structure, and they related the T1291C swap to the cause of hearing loss.
In the present study, it was evaluated the presence of the mitochondrial A1555G mutation in the MT-RNR1 (12S rRNA) gene in 70 hearing individuals, in Maringα and the regions. All participants showed normal pattern for the tested areas, that is, no positive cases or any polymorphisms were found for this mutation.
In the United States, in a neonatal trial, only one out of 1,173 subjects showed the A1555G mutation. [17] This mutation was not found in either 712 hearing subjects in a trial carried out in Argentina, [18] or in a neonatal trial conducted in Brazil with 100 children. [16]
In Brazil, Abreu-Silva et al., [15] did not find the A1555G mutation when analyzing 300 hearing subjects, among them 190 Afro-Brazilians and 110 Euro-Brazilians. In this study, there were a higher number of Euro-Brazilians (87.1%), against Afro-Brazilians (8.6%) and Nippon-Brazilians (4.3%). Although it was not possible to determine the prevalence of it among the different ethnicities, we can affirm that this mutation is not prevalent, or it must be very rare in hearing subjects in the State of Paranα, the south region of Brazil.
The frequency of A1555G mutation in the hearing-impaired people found in our study (1.3%) and as the cause of deafness is similar to those found in other Brazilian populations. Consequently, it should be examined in deafness diagnosis. The investigation of the A1555G mutation can contribute towards the determination of the nonsyndromic deafness etiology, hence, contributing to the correct genetic counseling process. This would justify its inclusion in routine exams for the etiology of nonsyndromic deafness, especially when there is a history of maternally inherited hearing impairment.
 References References | |  |
| 1. | Van Camp G, Smith R. Hereditary Hearing Loss Homepage, Last accessed June 20, 2012. Available at http://www.hereditaryhearingloss.org. 
|
| 2. | MITOMAP: A human mitochondrial genome database, Last accessed June 20, 2012. Available at http://mitomap.org/MITOMAP. 
|
| 3. | Van Camp G, Smith RJ. Maternally inherited hearing impairment. Clin Genet 2000;57:409-14. 
|
| 4. | Samanich J, Lowes C, Burk R, Shanske S, Lu J, Shanske A, et al. Mutations in GJB2, GJB6, and mitochondrial DNA are rare in African American and Caribbean Hispanic individuals with hearing impairment. Am J Med Genet A 2007;143A: 830-8. 
|
| 5. | Hamasaki K, Rando RR. Specific binding of aminoglycosides to a human rRNA construct based on a DNA polymorphism which causes aminoglycoside-induced deafness. Biochemistry 1997;36:12323-8. 
|
| 6. | Ballana E, Morales E, Rabionet R, Montserrat B, Ventayol M, Bravo O, et al. Mitochondrial 12S rRNA gene mutations affect RNA secondary structure and lead to variable penetrance in hearing impairment. Biochem Biophys Res Commun 2006;341:950-7. 
|
| 7. | Estivill X, Govea N, Barceló E, Badenas C, Romero E, Moral L, et al. Familial progressive sensorineural deafness is mainly due to the mtDNA A1555G mutation and is enhanced by treatment of aminoglycosides. Am J Hum Genet 1998;62:27-35. 
|
| 8. | Usami S, Abe S, Akita J, Shinkawa H, Kimberling WJ. Sensorineural hearing loss associated with the mitochondrial mutations. Adv Otorhinolaryngol 2000;56:203-11. 
|
| 9. | Kupka S, Tóth T, Wróbel M, Zeissler U, Szyfter W, Szyfter K, et al. Mutation A1555G in the 12S rRNA gene and its epidemiological importance in German, Hungarian, and Polish patients. Hum Mutat 2002;19:308-9. 
|
| 10. | Malik SG, Pieter N, Sudoyo H, Kadir A, Marzuki S. Prevalence of the mitochondrial DNA A1555G mutation in sensorineural deafness patients in island Southeast Asia. J Hum Genet 2003;48:480-3. 
|
| 11. | ØStergaard E, Montserrat-Sentis B, Grønskov K, Brøndum-Nielsen K. The A1555G mtDNA mutation in Danish hearing-impaired patients: Frequency and clinical signs. Clin Genet 2002;62:303-5. 
|
| 12. | Meza G, Torres-Ruíz NM, Tirado-Gutiérrez C, Aguilera P. mtDNA mutations, hearing loss and aminoglycoside treatment in Mexicans. Braz J Otorhinolaryngol 2011;77:573-6. 
|
| 13. | Khalifa Alkowari M, Girotto G, Abdulhadi K, Dipresa S, Siam R, Najjar N, et al. GJB2 and GJB6 genes and the A1555G mitochondrial mutation are only minor causes of nonsyndromic hearing loss in the Qatari population. Int J Audiol 2012;51:181-5. 
|
| 14. | Cecatto SB, Garcia RID, Costa KC, Abdo TR, Rezende CE, Rapoport PB. Análise das principais etiologias de deficiência auditiva em Escola Especial "Anne Sullivan". Braz JOtorhinolaryngol2003;69:235-40. 
|
| 15. | Abreu-Silva RS, Lezirovitz K, Braga MC, Spinelli M, Pirana S, Della-Rosa VA, et al. Prevalence of the A1555G (12S rRNA) and tRNASer (UCN) mitochondrial mutations in hearing-impaired Brazilian patients. Braz J Med Biol Res 2006;39:219-26. 
|
| 16. | Maniglia LP, Moreira BC, Silva MA, Piatto VB, Maniglia JV. Screening of the mitochondrial A1555G mutation in patients with sensorineural hearing loss. Braz J Otorhinolaryngol 2008;74:731-6. 
|
| 17. | Tang HY, Hutcheson E, Neill S, Drummond-Borg M, Speer M, Alford RL. Genetic susceptibility to aminoglycoside ototoxicity: How many are at risk? Genet Med 2002;4:336-45. 
|
| 18. | Gravina LP, Foncuberta ME, Estrada RC, Barreiro C, Chertkoff L. Carrier frequency of the 35delG and A1555G deafness mutations in the Argentinean population. Impact on the newborn hearing screening. Int J Pediatr Otorhinolaryngol 2007;71:639-43. 
|
[Figure 1]
[Table 1]
|






