|
 
 |
|
ORIGINAL ARTICLE |
|
|
|
| Year : 2013 | Volume
: 19
| Issue : 1 | Page : 58-64 |
| |
Association of interleukin-4 and interleukin-17F polymorphisms in periodontitis in Dravidian ethnicity
Nidhi Jain1, Rosamma Joseph1, Shabeesh Balan2, R Arun1, Moinak Banerjee2
1 Department of Periodontics, Government Dental College, Calicut, India
2 Department of Human Molecular Genetics, Rajiv Gandhi Centre for Biotechnology, Thiruvananthapuram, Kerala, India
| Date of Web Publication | 4-Jun-2013 |
Correspondence Address:
Rosamma Joseph
Department of Periodontics, Government Dental College, Medical College P.O, Calicut, Kerala
India
 Source of Support: None, Conflict of Interest: None  | 3 |
DOI: 10.4103/0971-6866.112891

 Abstract Abstract | | |
Background: Complex network of pro and anti-inflammatory cytokines are known to act in inflamed periodontal tissue. This study explores the distribution of interleukin (IL)-4 (+33 C/T) and IL-17F (7383A/G, 7488A/G) gene polymorphism in chronic and aggressive periodontitis subjects of Dravidian ethnicity.
Materials and Methods: This case control study consisted of 124 periodontitis individuals comprising of 63 chronic and 61 aggressive periodontitis subjects as cases, and control group consisted of 101 healthy subjects. All subjects were genotyped for IL-4 + 33C/T, IL-17F 7383A/G, 7488A/G by polymerase chain reaction amplification followed by TaqMan assay for IL-4 + 33C/T, restriction enzyme digestion and gel electrophoresis for IL-17F 7383A/G and sequencing for IL-17F 7488A/G.
Results: IL-4 + 33C/T was significantly associated with periodontitis (P < 0.05) at both allelic and genotypic level. In subgroup analysis also significant difference (P < 0.05) in allelic distribution between aggressive periodontitis and control group for loci IL-4 + 33C/T was noted. However, there was a lack of association between IL-17F 7383A/G and IL-17F 7488A/G with periodontitis and its sub-groups at both allelic and genotypic levels.
Conclusions: In Malayalam speaking Dravidian population IL-4 + 33C/T loci appears to be an important risk factor for periodontal disease with a leaning towards aggressive periodontitis. The association between IL-17F at 7383A/G and 7488A/G loci with either chronic or an aggressive periodontitis could not be ascertained.
Keywords: Aggressive periodontitis , cytokines, host response, periodontitis, polymorphism
How to cite this article:
Jain N, Joseph R, Balan S, Arun R, Banerjee M. Association of interleukin-4 and interleukin-17F polymorphisms in periodontitis in Dravidian ethnicity. Indian J Hum Genet 2013;19:58-64 |
How to cite this URL:
Jain N, Joseph R, Balan S, Arun R, Banerjee M. Association of interleukin-4 and interleukin-17F polymorphisms in periodontitis in Dravidian ethnicity. Indian J Hum Genet [serial online] 2013 [cited 2016 May 24];19:58-64. Available from: http://www.ijhg.com/text.asp?2013/19/1/58/112891 |
 Introduction Introduction | |  |
Periodontitis is a chronic and complex inflammatory disease caused by bacterial biofilm, characterized by progressive destruction of supporting structures of teeth. Although periodontal diseases are initiated by bacteria, the host response plays a crucial role in the breakdown of the connective tissue and bone. [1] The host response is influenced by several possible risk factors such as oral hygiene, smoking, age, gender, and genetic factors. [2]
Cytokines play a pivotal role in the regulation of immune response [3] T helper1 (Th1) subset cells secrete interferon gamma and mediate cellular immunity, Th2 subset secrete interleukin (IL)-4 and activates allergic and humoral response. [4] Th17, a novel subset of Th cells is distinct from Th1 to Th2 and selectively produce IL-17.
IL-4, a potent down regulator of macrophage function, inhibits the secretion of proinflammatory cytokines such as IL-1, IL-6, and tumor necrosis factor (TNFα). The recent findings indicate that periodontitis is associated with increased levels of IL-4 [5],[6] whereas conflicting reports are also available. [7],[8] These differences in observations can be attributed to the population differences in the rates of production of IL-4.Genetic factors could play a role in the IL-4 systems and IL-4 is a reasonable candidate gene for susceptibility to severity of chronic inflammatory conditions like periodontal diseases.
IL-17F is a novel proinflammatory cytokine that regulates inflammation, osteoclast activity, amplify proinflammatory response, and promotes autoimmunity. The role of IL-17 in periodontal diseases is poorly understood. [9] The available evidences suggest that IL-17F gene is an excellent candidate gene for chronic inflammatory disease like rheumatoid arthritis (RA). [10] RA and periodontal diseases share a common immuno-inflammatory pathway. Association between RA and chronic periodontitis has been well-documented in periodontal literature. [11],[12] No reports, until now are available in periodontal literature investigating an association between IL-17F gene polymorphism and periodontal diseases.
Genetic associations found in one population need not necessarily hold true in other population. There are no reports available in the literature on the distribution of IL-4 and IL-17F gene polymorphism in subjects of Dravidian ethnicity. Dravidian ethnicity includes Tamil, Telugu, Kannada, and Malayalam-speaking people. The objective of the study was to analyze the allelic and genotypic association of IL-4 + 33C/T, IL-17F 7383A/G, 7488A/G polymorphisms with periodontitis and its subgroups, the chronic periodontitis (CHP) and aggressive periodontitis (AP) in a Malayalam-speaking Dravidian population from South India.
 Materials and Methods Materials and Methods | |  |
Study design
This case control study was conducted from November 2010 to October 2011 among 225 subjects of Malayalam-speaking Dravidian ethnicity from South India. The study was carried out with a written informed consent from the subjects and was approved by the Institutional Ethics Committee, Dental College, Calicut, Kerala, India. The case group included a total of 124 subjects comprising of 63 chronic periodontitis and 61 aggressive periodontitis patients. Control group comprised of 101 subjects who were either periodontally healthy or having mild periodontitis. The mean age was 37.56 years (age range 19-50 years) in the group with the chronic periodontitis and 25.29 years (age range 13-40) in the group with an aggressive periodontitis.
Selection of study subjects
The study population was defined based on the criteria proposed by the American Academy of Periodontology [13] and the clinical case definitions proposed by the Division of Oral Health at the Centers for Disease Control and Prevention for use in population-based surveillance of periodontitis. [14] The case groups consisted of patients who attended the out-patient wing of the Department of Periodontics, Government Dental College, Calicut, Kerala, India who were of Dravidian ethnicity with Malayalam as their mother tongue. They were diagnosed as having either chronic periodontitis (CHP) or aggressive periodontitis (AP). Patients with history of systemic disease, bleeding disorder, immunosuppressive disease, tobacco users, pregnant females, and a history of prior periodontal therapy in last 6 months were excluded from the study.
The control group was selected from systemically healthy volunteers and among the staff and students from Government Dental College, Calicut, Kerala, India. The selected control samples were of the same ethnic origin.
Clinical examination
A complete clinical examination was carried out by a single trained examiner (NJ). A standardized periodontal probe with William's graduated markings was used. Measurements of probing depth, clinical attachment level, the plaque index, calculus index (calculus component of the Simplified Oral Hygiene Index), and gingival index were recorded.
Molecular methods
A peripheral blood sample (3 ml) was collected from all subjects by venipuncture from the antecubital fossa and transferred to plastic falcon tubes containing EDTA. A modified, standard, organic extraction method was used for DNA extraction as proposed by Sambrook et al. [15]
The IL-4 + 33C/T polymorphism, genotyping was carried out by TaqMan® genotyping assays as per manufacturer's protocol (Applied Biosystems, USA). The SNPs at position + 33C/T of the IL-4 gene were amplified using thermocycling conditions as follows: Initial denaturation at 95°C for 10 min followed by 40 cycles of 15 sec at 92°C (denaturation) and 1 min at 60°C (annealing/extension). Thermal cycling in 7900HT Real-Time polymerase chain reaction (PCR) System and data analysis was performed by the SDS2.1 Software for Applied Biosystems, USA.
The SNP's at position 7383A/G, and 7488A/G of IL-17F gene was amplified using Forward 5'- GTGTAGGAACTTGGGCTGCA-3' and Reverse primers 5'- AGCTGGGAATGCAAAC AAAC-3' under the thermocycling conditions: Initial denaturation at 95°C for 2 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 50°C for 30 s, extension at 72°C for 30 s, and final extension at 72°C for 5 min. The PCR products thus obtained for IL-17F 7383A/G were digested with 1 μl of AvaII at 37°C for overnight. AvaII restriction digestion of PCR product yielded 470 bp for allele A and 75 and 395 bp for allele G which were visualized on 3% agarose gel electrophoresis by ethidium bromide staining. The heterozygous individuals were identified by the presence of three bands.
The amplified product obtained for IL-17F 7488A/G was used as the template for the sequencing PCR as per the manufacturers protocol (Applied Biosystems, USA). The PCR product after the Cycle sequencing was cleaned up to remove the unbound nucleotides. The samples were denatured at 95°C for 3 min by addition of 10 μl of Hi-Di™ formamide and subsequently loaded into ABI3130XL Genetic Analyzer. The sequence data was analyzed using the Software Applied Biosystems Sequence Scanner v1.0.
Statistical analysis
The clinical variables between the cases and controls and within the subgroups of the case groups were compared using the one-way ANOVA test. The distribution of allelic and genotypic frequencies at IL-4 + 33C/T and IL-17F 7383A/G, 7488A/G among the different study group were also analyzed. Hardy-Weinberg equilibrium was tested for genotype frequencies. Statistical significance was set at P < 0.05. All the analyses were performed using a statistical package termed SPSS 13.
 Results Results | |  |
Baseline clinical parameters of case groups
The periodontal parameters of the study subjects are shown in [Table 1]. All the periodontal parameters were significantly different between the case and control group (One-way ANOVA, P < 0.001). After post hoc adjustment all the periodontal clinical parameters were found to be significantly higher in both case groups as compared to healthy controls. Calculus index scores were significantly lower in AP as compared to CHP group (P < 0.001).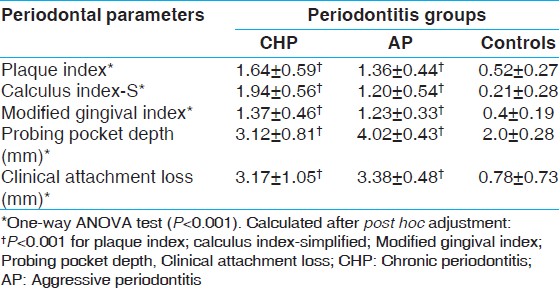 | Table 1: Comparison of periodontal parameters between the groups (mean±SD)
Click here to view |
Comparison of IL-4 + 33C/T and IL-17F 7383A/G and 7488A/G in cases and controls
The comparison of allelic and genotypic association of IL-4 + 33C/T polymorphism revealed a significant difference between the control group and overall periodontitis group (AP + CHP) [Table 2]. The C allele and CC homozygous genotype was over-represented in the periodontitis group. However, when the periodontitis groups (CHP or AP) were individually compared within and between the subgroups and the control group, no statistically significant difference in the genotype distribution of IL-4 + 33C/T was observed. Interestingly, a significant allelic association was observed with C allele for aggressive periodontitis against the control, in the sub-group analysis [Table 3] and [Table 4].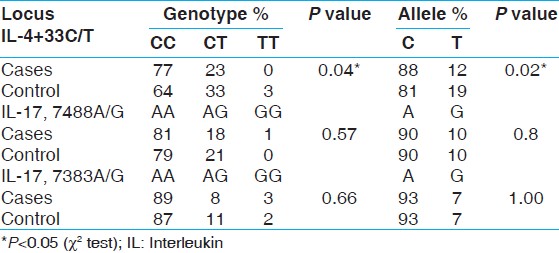 | Table 2: Allele and genotype frequencies of IL - 4+33, IL-17, 7488A/G, 7383A/G polymorphisms in periodontitis cases (aggressive and chronic periodontitis) and controls
Click here to view |
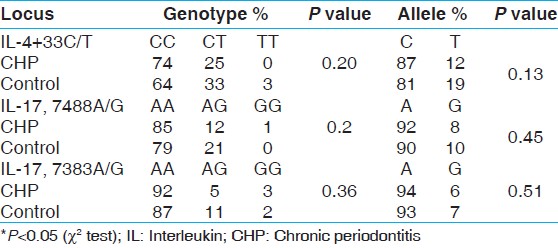 | Table 3: Allele and genotype frequencies of IL-4 + 33, IL-17, 7488A/G, 7383A/G in chronic periodontitis and controls
Click here to view |
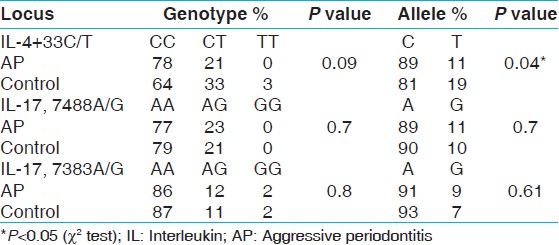 | Table 4: Allele and genotype frequencies of IL-4 + 33, IL-17, 7488A/G, 7383A/G polymorphisms in aggressive periodontitis and controls
Click here to view |
No significant difference was observed in genotype distribution and allelic frequencies of IL-17F at position 7383A/G and 7488A/G in overall group with periodontitis (CHP + AP) and controls [Table 2]. No allelic or genotypic associations were observed with IL-17F polymorphism with CHP or AP group was found in the present study [Table 3] and [Table 4].
 Discussion Discussion | |  |
Periodontitis is a chronic and complex inflammatory disease caused by bacterial biofilm and progression of diseases depends upon the production of host mediators. Complex network of pro and anti-inflammatory cytokines (Th1, Th2 and Th17) are known to act in inflamed periodontal tissue. Periodontitis has typically multiple gene associations and different allelic variants when combined with environmental factors can result in variable diseases manifestations. Genetic variations like polymorphisms in the genes coding for proinflammatory cytokines are potential candidates for susceptibility to periodontitis. Single Nucleotide gene polymorphism is likely to be valuable tools for genetic study to assess risk allele for a disease as they represent variation in the population.
IL-4 and IL-17 play an important role in the pathogenesis of periodontal diseases as anti-inflammatory cytokine and pro-inflammatory cytokine respectively. Hence studying the genetic polymorphisms in these two cytokine genes could be alluring. To our knowledge no studies have examined IL-4 and IL-17F gene polymorphism in the Indian subcontinent within subjects of Dravidian ethnicity. One of the major concerns related to genotype studies is the sample size of the study subjects. Our study population consisted of total 225 subjects. This sample size is relatively higher than many of studies reported earlier. [16],[17],[18],[19]
In the present study, we found statistically significant difference in distributions of genotype CC, CT, TT for IL-4 + 33C/T among cases (CHP + AP) and the control group. The distribution of genotype CC (77%) was higher in case group as compared to controls (64%). This distribution of genotype is in accordance with the study carried out in an Iranian population [20] where they found predominance of CC genotype in cases (CHP + AP), however, they did not attain the statistical significance.
However, when the periodontal diseases were stratified into aggressive and chronic periodontitis, we do find a statistical significant difference in allelic distribution of SNP IL-4 + 33C/T between aggressive periodontitis (C = 89%, T = 11%) and control group (C = 81%, T = 19%) but not with genotypes. In contrast to our observation, no significant differences in allelic distribution between aggressive periodontitis patients and controls were found in Caucasian and Japanese population. [21] They also found the prevalence of T allele to be 38% in AP and 33% in healthy control group, which is reverse of our findings, where T allele frequency is higher in control group (19%) as compared to the AP group (5.88%). Interestingly, the same authors when stratified the study purely based on caucasian ethnicity, they reported a statistically significant difference in IL-4 genotypes and aggressive periodontitis [22] . Similar findings had been reported by Michel (2001) in Caucasian population. [23] He found 27.8% of subjects with early onset periodontitis were IL-4 promoter and intron polymorphism positive (PP + and IP+) and none of the healthy controls presented a polymorphism, indicating a specificity of 100%. He also quantified the serum levels of IL-4 in patients who were positive for both polymorphisms (PP + and IP+). However, he was not able to quantify levels of IL-4 in the sera of the PP + and IP + patients by ELISA as they were below the limit of detection. In an earlier study we reported the predominance of C allele of IL-1β+3954 polymorphism in AP cases in the Dravidian population. [24]
In this study, we found no statistically significant association between chronic periodontitis group and controls for SNP IL-4 + 33C/T for genotypic distribution and allelic frequency. It is consistent with the findings in different population such as Korean, [25] Brazilian, [26] Turkish, [27] Iranian, [20] and Czech origin [28] in which they concluded that IL4 gene is not associated with the susceptibility to chronic periodontal disease. The C allele frequency of IL-4 + 33C/T in chronic periodontitis (87.3%) in our study was higher than in control group (81%), however, there is no statistically significant difference. Similar lack of association for IL-4 + 33C/T was reported in Brazilian population where they reported no significant difference in genotype and allele frequencies between chronic periodontitis and control group but C allele was more predominant in subjects with chronic periodontitis [26] . However, contrasting results were also reported but in Czech patients where they found higher frequency of C allele in control group as compared to chronic periodontitis [28] . From this study, it can be inferred that IL-4 gene polymorphism preferably the C allele could have influenced the pathogenesis of periodontal diseases (CHP + AP) in Dravidian population with a leaning towards aggressive periodontitis rather than chronic periodontitis.
Although IL-4 genetically determines the differences in the cytokine expression levels, bacteria such as P.gingivalis  and Fusobacterium nucleatum can also interact with it and could form a higher level of IL-4. As IL-4 levels have not been detected in the present study, larger sample size, which evaluates IL-4 levels will be more explanatory regarding the role of IL-4 polymorphism in CHP. Furthermore, the promoter of the IL-4 gene largely contributes to the control of T-cell differentiation, as a key binding site for specific Th-2 transcription factors. Therefore, along with IL-4 + 33C/T, IL-4 promoter polymorphism, and IL4 levels would provide more informative observation regarding the association between IL-4 and chronic periodontitis. Although in the current study no association could be demonstrated between IL-4 genotypes and chronic periodontitis, we cannot exclude that other polymorphic variation present within these genes, which may denote susceptibility to periodontal diseases. and Fusobacterium nucleatum can also interact with it and could form a higher level of IL-4. As IL-4 levels have not been detected in the present study, larger sample size, which evaluates IL-4 levels will be more explanatory regarding the role of IL-4 polymorphism in CHP. Furthermore, the promoter of the IL-4 gene largely contributes to the control of T-cell differentiation, as a key binding site for specific Th-2 transcription factors. Therefore, along with IL-4 + 33C/T, IL-4 promoter polymorphism, and IL4 levels would provide more informative observation regarding the association between IL-4 and chronic periodontitis. Although in the current study no association could be demonstrated between IL-4 genotypes and chronic periodontitis, we cannot exclude that other polymorphic variation present within these genes, which may denote susceptibility to periodontal diseases.
In our study, there was no statistically significant association between cases (CHP + AP) and the control group for both genotype and allele frequency of IL-17F at 7488A/G and 7383A/G polymorphisms. Even the sub-group analysis also did not reveal any signification association with IL-17F at 7488A/G and 7383A/G polymorphisms. No study is available in the periodontal literature until date, has investigated the influence of IL-17F polymorphisms in subjects with periodontitis. Rheumatoid arthritis (RA) and chronic periodontitis are the most common chronic inflammatory diseases with remarkable pathological and clinical similarities. In an earlier study the association between IL-17F gene polymorphisms and susceptibility to and severity of RA in Polish patients [10] have been investigated and their results were in accordance with our study, suggesting no evidence of association of IL17F. The authors in an earlier study reported that occurrence and severity of periodontitis was found to be higher in RA subjects as compared to subjects without RA, suggesting a positive relation between these two chronic inflammatory diseases. [12] The G allele frequency of IL-17F in our control group was higher than that of chronic periodontitis group, whereas in Polish subjects higher frequency of G allele was reported in RA cases. [10] In ulcerative colitis patients homozygous polymorphic (GG) genotype of IL-17 was lower than in healthy subjects. [29] In the present study, the frequency of homozygous polymorphic genotype (GG) is almost similar in both chronic periodontitis and control groups.Even though, we did not observe a statistically significant association between IL-17F gene polymorphism and periodontal diseases, we could not exclude other polymorphic variation present within this gene and interaction with periodontal pathogen may also be involved. To know the exact relationship between the IL-17F gene polymorphism and periodontal diseases, further studies associated with IL-17F expression and its genetic analysis in large periodontitis cohorts with clinical data is warranted.
The limitations of our study are that the level of IL-4 and IL-17 has not been assessed in gingival crevicular fluid or periodontal tissues in study population. Furthermore, a large sample size would have provided better significance. No correlation was carried out for etiologic factors i.e., the plaque or calculus indices with genotypes of the study subjects.
 Conclusions Conclusions | |  |
In Malayalam speaking Dravidian population, CC genotype as well as allele C of IL-4 + 33C/T loci appears to be an important risk factor for periodontal disease (CHP + AP) with a leaning towards aggressive periodontitis. We did not find any association with chronic periodontitis at IL-4 + 33C/T loci. We do not associate the SNPs of IL-17F 7383A/G and 7488A/G to play any significant role in either chronic periodontitis or aggressive periodontitis.
 References References | |  |
| 1. | Kornman KS, Knobelman C, Wang HY. Is periodontitis genetic? The answer may be Yes! J Mass Dent Soc 2000; 49:26-30. 
|
| 2. | Kornman KS, di Giovine FS. Genetic variations in cytokine expression: A risk factor for severity of adult periodontitis. Ann Periodontol 1998;3:327-38. 
|
| 3. | Kinane DF, Hart TC. Genes and gene polymorphisms associated with periodontal disease. Crit Rev Oral Biol Med 2003;14:430-49. 
|
| 4. | Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol 1986;136:2348-57. 
|
| 5. | McFarlane CG, Meikle MC. Interleukin-2, interleukin-2 receptor and interleukin-4 levels are elevated in the sera of patients with periodontal disease. J Periodontal Res 1991;26:402-8. 
|
| 6. | Yamazaki K, Nakajima T, Gemmell E, Polak B, Seymour GJ, Hara K. IL-4-and IL-6-producing cells in human periodontal disease tissue. J Oral Pathol Med 1994;23:347-53. 
|
| 7. | Giannopoulou C, Kamma JJ, Mombelli A. Effect of inflammation, smoking and stress on gingival crevicular fluid cytokine level. J Clin Periodontol 2003;30:145-53. 
|
| 8. | Pradeep AR, Roopa Y, Swati PP. Interleukin-4, a T-helper 2 cell cytokine, is associated with the remission of periodontal disease. J Periodontal Res 2008;43:712-6. 
|
| 9. | Oda T, Yoshie H, Yamazaki K. Porphyromonas gingivalis antigen preferentially stimulates T cells to express IL-17 but not receptor activator of NF-kappaB ligand in vitro. Oral Microbiol Immunol 2003;18:30-6. 
|
| 10. | Paradowska-Gorycka A, Wojtecka-Lukasik E, Trefler J, Wojciechowska B, Lacki JK, Maslinski S. Association between IL-17F gene polymorphisms and susceptibility to and severity of rheumatoid arthritis (RA). Scand J Immunol 2010;72:134-41. 
|
| 11. | Farquharson D, Butcher JP, Culshaw S. Periodontitis, Porphyromonas, and the pathogenesis of rheumatoid arthritis. Mucosal Immunol 2012;5:112-20. 
|
| 12. | Joseph R, Rajappan S, Nath SG, Paul BJ. Association between chronic periodontitis and rheumatoid arthritis: A hospital-based case-control study. Rheumatology international 2013, 33, 103-9. 
|
| 13. | Armitage GC. Development of a classification system for periodontal diseases and conditions. Ann Periodontol 1999;4:1-6. 
|
| 14. | Page RC, Eke PI. Case definitions for use in population-based surveillance of periodontitis. J Periodontol 2007;78:1387-99. 
|
| 15. | Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. Vol. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. 
|
| 16. | Kornman KS, Crane A, Wang HY, di Giovine FS, Newman MG, Pirk FW, et al. The interleukin-1 genotype as a severity factor in adult periodontal disease. J Clin Periodontol 1997;24:72-7. 
|
| 17. | Galbraith GM, Hendley TM, Sanders JJ, Palesch Y, Pandey JP. Polymorphic cytokine genotypes as markers of disease severity in adult periodontitis. J Clin Periodontol 1999;26:705-9. 
|
| 18. | McGuire MK, Nunn ME. Prognosis versus actual outcome. IV. The effectiveness of clinical parameters and IL-1 genotype in accurately predicting prognoses and tooth survival. J Periodontol 1999;70:49-56. 
|
| 19. | Mark LL, Haffajee AD, Socransky SS, Kent RL Jr, Guerrero D, Kornman K, et al. Effect of the interleukin-1 genotype on monocyte IL-1beta expression in subjects with adult periodontitis. J Periodontal Res 2000;35:172-7. 
|
| 20. | Hooshmand B, Hajilooi M, Rafiei A, Mani-Kashani KH, Ghasemi R. Interleukin-4 (C-590T) and interferon-gamma (G5644A) gene polymorphisms in patients with periodontitis. J Periodontal Res 2008;43:111-5. 
|
| 21. | Gonzales JR, Kobayashi T, Michel J, Mann M, Yoshie H, Meyle J. Interleukin-4 gene polymorphisms in Japanese and Caucasian patients with aggressive periodontitis. J Clin Periodontol 2004;31:384-9. 
|
| 22. | Gonzales JR, Mann M, Stelzig J, Bödeker RH, Meyle J. Single-nucleotide polymorphisms in the IL-4 and IL-13 promoter region in aggressive periodontitis. J Clin Periodontol 2007;34:473-9. 
|
| 23. | Michel J, Gonzáles JR, Wunderlich D, Diete A, Herrmann JM, Meyle J. Interleukin-4 polymorphisms in early onset periodontitis. J Clin Periodontol 2001;28:483-8. 
|
| 24. | Shete AR, Joseph R, Vijayan NN, Srinivas L, Banerjee M. Association of single nucleotide gene polymorphism at interleukin-1beta+3954,-511, and-31 in chronic periodontitis and aggressive periodontitis in Dravidian ethnicity. J Periodontol 2010;81:62-9. 
|
| 25. | Kang BY, Choi YK, Choi WH, Kim KT, Choi SS, Kim K, et al. Two polymorphisms of interleukin-4 gene in Korean adult periodontitis. Arch Pharm Res 2003;26:482-6. 
|
| 26. | Scarel-Caminaga RM, Trevilatto PC, Souza AP, Brito RB Jr, Line SR. Investigation of IL4 gene polymorphism in individuals with different levels of chronic periodontitis in a Brazilian population. J Clin Periodontol 2003;30:341-5. 
|
| 27. | Kara N, Keles GC, Sumer P, Gunes SO, Bagci H, Koprulu H, et al. Association of the polymorphisms in promoter and intron regions of the interleukin-4 gene with chronic periodontitis in a Turkish population. Acta Odontol Scand 2007;65:292-7. 
|
| 28. | Holla LI, Fassmann A, Augustin P, Halabala T, Znojil V, Vanek J. The association of interleukin-4 haplotypes with chronic periodontitis in a Czech population. J Periodontol 2008;79:1927-33. 
|
| 29. | Chen B, Zeng Z, Hou J, Chen M, Gao X, Hu P. Association of interleukin-17F 7488 single nucleotide polymorphism and inflammatory bowel disease in the Chinese population. Scand J Gastroenterol 2009;44:720-6. 
|
[Table 1], [Table 2], [Table 3], [Table 4]
|






