|
 
 |
|
ORIGINAL ARTICLE |
|
|
|
| Year : 2013 | Volume
: 19
| Issue : 1 | Page : 65-70 |
| |
Distribution of CC-chemokine receptor-5-∆32 allele among the tribal and caste population of Vidarbha region of Maharashtra state
Arvind B Chavhan1, Santosh S Pawar2, Rajusing G Jadhao3, Kishor G Patil4
1 Department of Zoology, Shri Shivaji Science College, Amravati; Institute of Science, Nagpur, India
2 Department of Zoology, Govt. Vidarbha Institute of Science and Humanities, Amravati, Maharashtra, India
3 Department of Zoology, Shri Shivaji Science College, Amravati, India
4 Department of Zoology, Institute of Science, Nagpur, India
| Date of Web Publication | 4-Jun-2013 |
Correspondence Address:
Arvind B Chavhan
Department of Zoology, Shri Shivaji Science College, Morshi Road, Amravati, Maharashtra
India
 Source of Support: None, Conflict of Interest: None
DOI: 10.4103/0971-6866.112894

 Abstract Abstract | | |
Background: Genetic relationships among the ethnic groups are not uniform across the geographical region. Considering this assumption, we analyzed the frequency of the CC-chemokine receptor-5 (CCR5)-∆32 allele of the CCR5 chemokine receptor, which is considered a Caucasian marker, in Bhil tribal and Brahmin caste sample sets from the population.
Materials and Methods: 108 blood samples were collected from 6 tribe's populations and a caste population from the district of Vidarbha region.
Results and Discussion: The presence of low frequencies of CCR5-Δ32 in an individual of Bhil tribe (0.034, χ2 value 0.017) in the present study implies that these communities may have a better resistance toward human immunodeficiency virus (HIV)/acquired immunodeficiency syndrome (AIDS) than the other studied tribe sample, as non-show such mutation.
Conclusion: The marginal presence of the allele seen in the studied tribal population could be due to gene flow from the people of European descent. However, lack of the homozygous CCR5-Δ32 mutation and the low prevalence of heterozygous CCR5-Δ32 mutations suggest that the Indians are highly susceptible to HIV/AIDS, and this correlates with the highest number of HIV/AIDS infected individuals in India.
Keywords: Allele frequency, CC-chemokine receptor-5-∆32, India, genetic polymorphism, tribes, Vidarbha
How to cite this article:
Chavhan AB, Pawar SS, Jadhao RG, Patil KG. Distribution of CC-chemokine receptor-5-∆32 allele among the tribal and caste population of Vidarbha region of Maharashtra state. Indian J Hum Genet 2013;19:65-70 |
How to cite this URL:
Chavhan AB, Pawar SS, Jadhao RG, Patil KG. Distribution of CC-chemokine receptor-5-∆32 allele among the tribal and caste population of Vidarbha region of Maharashtra state. Indian J Hum Genet [serial online] 2013 [cited 2016 May 24];19:65-70. Available from: http://www.ijhg.com/text.asp?2013/19/1/65/112894 |
 Introduction Introduction | |  |
Human immunodeficiency virus (HIV)-1 infection has spread to all population groups in India and has reached epidemic proportions. [1] The rate of progression of HIV-1 disease exhibits a remarkable variation among different individuals. Many host genetic factors are now known to affect the disease progression rates, especially polymorphisms in genes encoding chemokine receptors. [2],[3],[4],[5] Although, no studies on chemokine receptor polymorphisms have been reported in the endogamous population of State of Maharashtra state so far, only one study has been carried out in healthy individuals of tribes and Muslim ethnic groups of Andhra Pradesh, south India. [6]
Certain members of the chemokine family of receptors serve as critical portals for the entry of HIV-1 into target cells. A mutant allele (CC-chemokine receptor-5 [CCR5]-∆32) of the β-chemokine receptor gene CCR5 carrying a 32 base-pair deletion prevents cell invasion by the primary transmitting strain of HIV-1. Individuals who are homozygous for the CCR5-∆32/∆32 allele are highly resistant to HIV-1 infection; the heterozygote state does not protect against HIV-1 infection, although, heterozygotes have been found to have significantly lower viral loads. [7] Early reports indicated that the CCR5-∆32 allele maybe absent in indigenous non-European populations. The CCR5_32 allele appears to have originated quite recently (approximately 7,000 years ago) in northeastern Europe. [8] Although its frequency has now reached a relatively high level in Europeans, e.g., 16.3% in Finns and 15.8% in Moravians, it is not present among African populations, and is only so at low levels in the Asian. [9] Hence, by application it is possible to evaluate the influence of the European population on the genetic constitution of others.
In India population one study has been carried out in healthy individuals of the tribes and Muslim ethnic groups of Andhra Pradesh, South India. [6] Majumder and Dey, [10] reported absence of CCR5-∆32 in various ethnic populations of India, both tribal and non-tribal, except for some populations of the northern and western regions where this allele may have been introduced by Caucasian gene flow. Although a few studies on chemokine, chemokine receptor, and DCSIGN exon 4 repeat number polymorphisms have been reported in north Indian (Aryan descent) HIV patients and healthy controls, [11],[12],[13] there is a dearth of reports on the HIV patients with and without tuberculosis (TB) of South Indian (Dravidian descent) origin.
CCR5-Δ32 exhibit variable frequencies in distinct populations [14],[15],[16],[17] and possibly, their phenotypes depend on the ethnicity analyzed. [15],[16],[17],[18],[19],[20] The Brazilian population presents a complex genetic background, characterized by a high degree of miscegenation. [21],[22] In major Brazilian cities, CCR5-Δ32 was found at frequencies of between 2% and 7%. [23],[24],[25],[26]
Genetic relationships among caste groups are not uniform across the geographical region of India . [27],[28] India is known for the enormous cultural and genetic diversity of its people. [27] Such diversity is some time attributed to the positioning of the Indian peninsula at the tri-junction of the three continents, viz. Africa, Europe, and Asia. The contemporary Indian population is stratified as tribal and non-tribal, i.e., caste population. The origin of the caste in India is an enigma, [29] though many are known to have a tribal origin. [30],[31]
The Maharashtra state of India forms a huge irregular triangle with its base on the west cost of India, overlooking the Arabian Sea. Historically, the state is comprised of three sub-regions, Western Maharashtra, the Vidarbha, and the Marathwada. Vidarbha lies on the eastern side and thus mainly contributes to the region broadly referred to as central India. Apart from the tribal population, many other Ethnic Communities mainly Hindus, Muslim, Buddhist and Sikhs, inhabit the region. The Vidarbha has a hoary past and has been under the domination of many Hindus, Muslims, and tribal-Gond Kingdoms. The Vidarbhian strip served as a bridge between Northern and SouthernIndia. It is assumed that the relationship between these various populations may define the present genetic landscape of India.
Taking this assumption and geographical and ethnic diversity into account in the present study, we investigated the distribution of CCR5-∆32 alleles in tribes from the Vidarbha region (Maharashtra) Central India.
 Material and Methods Material and Methods | |  |
Population
The Kolam (tribes)
Besides inhabiting the adjoining state, a substantial number of these people inhabit in few district of Vidarbha. They speak the Gondi dialect which belongs to the Dravidian linguistic group. We sampled 15 samples from this group Village of Yavatmal district.
The Bhil (tribes)
Bhils are listed as Adivasi residents of the states of Gujarat, Madhya Pradesh, Chhattisgarh, Maharashtra, and Rajasthan in Western and Central India as well as in Tripura in far-eastern India on the border with Bangladesh. Bhils are divided into a number of endogamous territorial divisions, which in turn have a number of clans and lineages. Most Bhils now speak the language of the region they reside in, such as Marathi and Gujarati. We sampled 15 samples from this group Village of Yavatmal district.
The Korkus (tribes)
The Korkus are a typical tribal population from Amravati district and found only in the Satpuda mountain ranges spanning Maharashtra and Madhya Pradesh. They are mainly concentrated in Melghat a scheduled area of Korku comprising 89% of the tribal population. The Korkus speak Korku dialect belonging to Austro-Asiatic linguistic group. The Austro-Asiatic speakers are considered as the first settler of Indian subcontinent. [32] We sampled 15 samples from this group.
The Paradhi (tribes)
Phase Paradhi or Phasse Paradhi is a tribe in India. The tribe often faces harassment by Indian law enforcement agencies. The tribe is found mostly in Maharashtra and parts of Madhya Pradesh. The Phasse are a sub tribe of the Paradhi caste, which includes sub-castes like Gav Paradhi, Berad-Paradhi, Gay-Paradhi, Chita Paradhi. Paradhi is the term for "hunter." There are only three surnames among them, Chauhan, Pawar, and Solanke. We sampled 15 samples from this group from Akola district.
The Andh (tribes)
A low cultivating caste of Berar, who numbered 52,000 persons in 1911, and belongs to the Yeotmal, Akola, and Buldana Districts. The Andhs appear to be a non-Aryan tribe of the Andhra or Tamil country, from which they derive their name. There were 8228 Andh in Andhra Pradesh in 1991. According to Singh et al.,[33] in their 2004 book people of India there are over 74,000 Andhs in Maharashtra. We sampled 16 samples from this group.
The Gonds (tribes)
This tribe falls under the primitive tribes category and spread much over the central India. Gond generally speaks "Gondi 0" dialect, which belongs to the Dravidian linguistic family, after Indo-European, in India. We sampled 17 blood samples from this tribe from the Village Gadchiroli district.
Brahmin caste
Brahmin is a class of priests and preachers of 'Dharma' and considered as the torch bearer of Hinduism. Majority of Brahmin in Maharashtra speak Marathi, one of the major languages of Indo-Aryan linguistic group. The population of Brahmins in Amravati district is 21,500 or 3% of the population. We collected 15 samples of Brahmins living in Amravati district.
Blood sample collection and DNA extraction
We have collected 108 blood samples on Whatman FTA mini cards (GE Health-Care, UK, Ltd) from 6 tribe's populations and a caste population from the district of Vidarbha region. Every card was labeled with appropriate code as per tribes/caste and district with informed consent obtained from each volunteer. Approximately, (200 μl [2-5 drop]) of blood by veni-puncture was directly spotted on the FTA mini card within a printed circle area and dried at room temperature. Genomic DNA was isolated from each dried blood sample by following protocol. [34]
Amplification and restriction digestion of DNA
Polymerase chain reaction was performed following previously prescribed protocol by. [35] Briefly, 100 ng of genomic DNA was denatured at 94°C for 10 min, following, which it was subjected to 30 cycles of denaturation, annealing, and extension. The last cycle was followed by an incubation at 72°C for 10 min. The reaction mixture of 50 μl contained, 50 mmol KCl, 10 mmol Tris-HCl, pH 8.3, 800 μmol dNTPs, 100 μg/ml gelatin, 10 pmoles of each of the CCR5-specific primer forward: CCR5 - F: (5'-CCTGGCTGTCGTCCATGCTG-3') and reverse: CCR5-R: (5'-CTGATCTAGAGCCATGTGCAC AACTCT-3')., and 1.5 units of Taq polymerase enzyme (Xcelris Genomics, Ahmedabad, India).
Genotyping for CCR5 Δ-32 polymorphism
The genotypes were visualized by running digested product on 2% agarose gel at 100 V for about 2 h and the results were recorded in gel documentation system. The EcoRI Restriction enzymes digest the amplified polymerization Chain Reaction (PCR) product of 735 base pairs (bps). The amplified product was digested with 10 U of EcoRI at 37°C for 2 h. After digestion, the products were analyzed on a 2% agarose gel and bands were visualized on a Ultraviolet (UV)-transilluminator. PCR amplified a 735 bp region of genomic DNA that spanned a 32-bp deletion differentiating the CCR5-_32 allele from its wild-type counterparts at the CCR5 locus. After restriction digestion with EcoRI, with wild gene yielded band at 332 bp and for mutated gene, the bands were at 332 and 403 [Figure 1].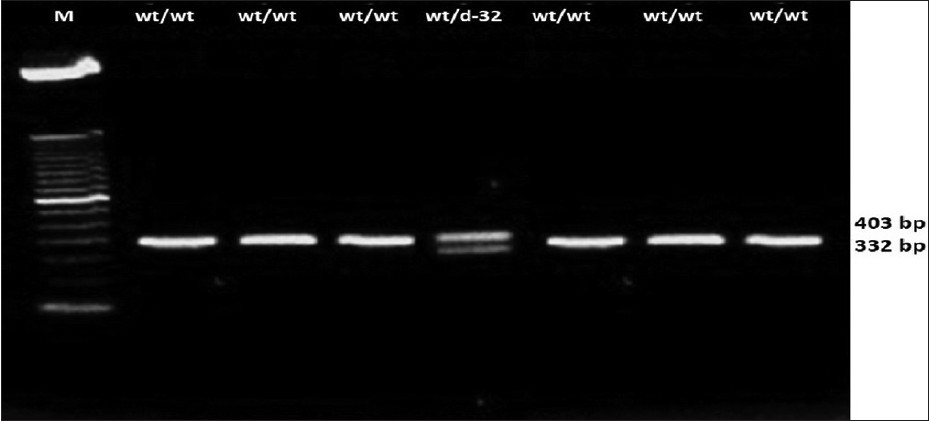 | Figure 1: CC - chemokine receptor - 5 genotyping among the tribes Lane 1, 2, 3, 5, 6 and 7 represent the PCR product from samples with homozygous wild type genotypes (fragments of 332 bp wt/wt). Lane 4 represents the Δ- 32 genotype (with the presence of both fragment of 332 and 403 wt/Δ32)
Click here to view |
Statistical analysis
Statistical analysis of allele frequencies was performed using Chi-square statistics. Genotype distribution for polymorphism was first compared to predictable values from Hardy-Weinberg equilibrium. In all cases, P values less than 0.05 were considered to be statistically significant.
 Observation and Results Observation and Results | |  |
Our data on distribution of CCR5-∆32 mutation among the selected tribe and a caste is depleted [Table 1]. Genotype and phenotype for the heterozygous mutation among the tribes sample suggested that it is either absent or present at low frequency 1.08% (1 in 93 tribe's samples and 1in 15 samples of a caste). None of the tribes and control caste was found to be homozygous for the CCR5-∆32 mutation, while the Bhil tribe and control caste show the heterozygous for the CCR5-∆32 mutation in negligible frequency (0.034). The analysis of χ2 suggested that the prevalence of the CCR5-∆32 is significantly low (χ2 = 0.02, P > 0.05). The aggregate frequencies of the entire sample for the wild-type allele CCR5 and the CCR-5-∆32 variant were found to be 0.991 and 0.009, respectively. Among the tribes Kolam, Korku, Paradhi, Andh and Gonds showed the highest (wild type) homozygous genotype frequency (100%). while only the Bhil tribe shows heterozygous genotype frequency (06.97%) and allelic frequency (0.034) for CCR5/∆32. Only one bearer of this mutation was found in the blood sample of Bhil tribe collected from Yavatmal district of Vidarbha region.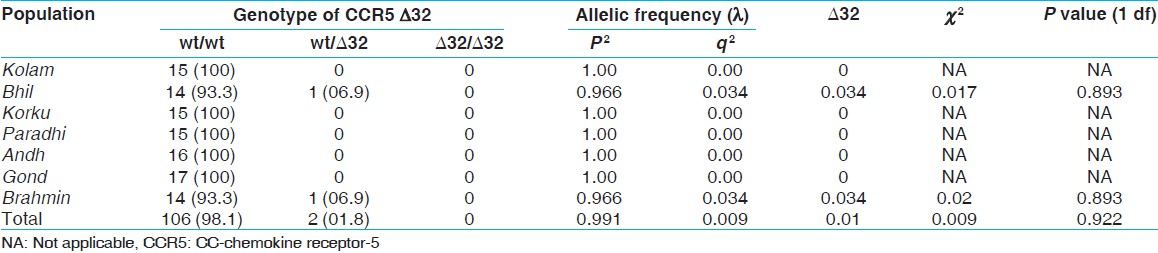 | Table 1: Genotypic distribution and gene frequencies of the CCR5 allele in different population samples of Vidarbha region of Maharashtra state
Click here to view |
No significant deviations from the HWE were observed (P > 0.05, Chi-squared goodness of fit). [Table 1] shows the frequency of the CCR5-∆32 allele in the six tribes and one control caste Brahmin population.
 Discussion on Conclusion Discussion on Conclusion | |  |
This study describes the genotype and allele frequencies of the polymorphisms CCR5-Δ32 in selected six tribal and one caste populations from Vidarbha region. Most importantly, however, this study is the first to be conducted in Vidarbha that investigates the genetic polymorphisms CCR5-∆32 among different ethnic tribal population settlement in the districts of Vidarbha region.
CCR5, a coreceptor for HIV-I virus, has been shown to be the most important for the HIV transmission . [36],[37] A 32-nucleotide deletion of CCR5 homozygous (CCR5-∆32/∆32) display a high degree of the natural resistance to HIV transmission whereas CCR5-∆32 heterozygosity (CCR5+/∆32) demonstrate a slower progression to acquired immunodeficiency syndrome (AIDS) than CCR5 wild type (CCR5+/+). [3],[38] However, this genetic mutation is found in Caucasian rather than non-Caucasian population including India. [10] The CCR5-∆32 genotype frequency among our study tribes sample was absent or negligible, the average genotype frequency were common homozygous wt/wt (98.3%), heterozygous wt/mt (1.87%) and rare homozygous mt/mt (0%), from the control group, revealing CCR5-∆32 allele frequency of 6.97%. The frequency of the CCR5-∆32 allele among our study population seems to be remarkably similar to previously reported frequencies in other Asian populations.
The CCR5-∆32 allele frequency among Asians is very low in Rajasthan Indians (0.05%), Andhra Pradesh Indians (0-0.03%), [39] North Indians (1.5%), [12] South Indians (1-3%), [6] and ethnic population of Kashmir (3-4%) . [40] A similar study conducted from the Island of Crete, Greece showed allele frequency of 3.25%, with a 95% confidence interval (CI) for conformity with Hardy-Weinberg equilibrium of 0.74-5.7%. [41] The CCR5-∆32 polymorphism is found all across Europe at different allele frequencies, with a North to South decreasing gradient and lower distribution in the regions of Southeast Mediterranean. [42] The frequency of the CCR5-∆32 allele in the studied tribal population is consistently similar with data reported from other populations with non-European ancestors. [9],[16] CCR5-Δ32 allelic frequencies were not different when the self-reported racial characteristics of the individuals evaluated were considered. [25] This allele has not been found among South American native Indians, corroborating the hypothesis of a European origin of this allele and its introduction to the continent through migration. [43]
Within the Middle-Eastern populations the frequency of the mutant CCR5-∆32 allele reached it's the highest among Iranians, 2.4%; Saudi, 2.1%; and it's the lowest among Kuwaitis, 1%; and the Egyptians, 0.5%; and is completely absent in individuals from the United Arab Emirates. [44] Our results suggest that the CCR5-∆32 allele is detected at very low frequency in studied tribal populations from Vidarbha. The presence of low frequencies of CCR5-Δ32 in an individual of Bhil tribe (0.034, χ2 value 0.017) in the present study implies that these communities may have a better resistance to HIV/AIDS than other studied tribe sample, as non show such mutation. However, the CCR5-∆32 allele is observed mostly in European populations.
The marginal presence of the allele seen in the studied tribal population could be due to gene flow from the people of European descent. However, CCR5-∆32 is completely absent in the populations from Africa, Oceania, and the Americas. However, lack of the homozygous CCR5-Δ32 mutation and the low prevalence of heterozygous CCR5-Δ32 mutations suggest that the Indians are highly susceptible to HIV/AIDS, and this correlates with the highest number of HIV/AIDS infected individuals in India.
 References References | |  |
| 1. | Misra SN, Sengupta D, Satpathy SK. AIDS in India: Recent trends in opportunistic infections. Southeast Asian J Trop Med Public Health 1998;29:373-6. 
|
| 2. | Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: Roles in viral entry, tropism, and disease. Annu Rev Immunol 1999;17:657-700. 
|
| 3. | O'Brien TR, Goedert JJ. Chemokine receptors and genetic variability: Another leap in HIV research. JAMA 1998;279:317-8. 
|
| 4. | Moore JP. Coreceptors: Implications for HIV pathogenesis and therapy. Science 1997;276:51-2. 
|
| 5. | Fauci AS. Host factors and the pathogenesis of HIV-induced disease. Nature 1996;384:529-34. 
|
| 6. | Ramana GV, Vasanthi A, Khaja M, Su B, Govindaiah V, Jin L, et al. Distribution of HIV-1 resistance-conferring polymorphic alleles SDF-1-3'A, CCR2-64I and CCR5-Delta32 in diverse populations of Andhra Pradesh, South India. J Genet 2001;80:137-40. 
|
| 7. | Huang Y, Paxton WA, Wolinsky SM, Neumann AU, Zhang L, He T, et al. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med 1996;2:1240-3. 
|
| 8. | Lidén K, Linderholm A, Götherström A. Pushing it back. Dating the CCR5-D32 bp deletion to the mesolithic in Sweden and its implications for the Meso/Neo transition. Doc Praehist 2006;33:29-37. 
|
| 9. | Martinson JJ, Chapman NH, Rees DC, Liu YT, Clegg JB. Global distribution of the CCR5 gene 32-basepair deletion. Nat Genet 1997;16:100-3. 
|
| 10. | Majumder PP, Dey B. Absence of the HIV-1 protective Delta ccr5 allele in most ethnic populations of India. Eur J Hum Genet 2001;9:794-6. 
|
| 11. | Suresh P, Wanchu A, Sachdeva RK, Bhatnagar A. Gene polymorphisms in CCR5, CCR2, CX3CR1, SDF-1 and RANTES in exposed but uninfected partners of HIV-1 infected individuals in North India. J Clin Immunol 2006;26:476-84. 
|
| 12. | Verma R, Gupta RB, Singh K, Bhasin R, Anand Shukla A, Chauhan SS, et al. Distribution of CCR5delta32, CCR2-64I and SDF1-3'A and plasma levels of SDF-1 in HIV-1 seronegative North Indians. J Clin Virol 2007;38:198-203. 
|
| 13. | Rathore A, Chatterjee A, Sivarama P, Yamamoto N, Singhal PK, Dhole TN. Association of RANTES-403 G/A,-28 C/G and In1.1 T/C polymorphism with HIV-1 transmission and progression among North Indians. J Med Virol 2008;80:1133-41. 
|
| 14. | Salem AH, Farid E, Fadel R, Abu-Hijleh M, Almawi W, Han K, et al. Distribution of four HIV type 1-resistance polymorphisms (CCR5-Delta32, CCR5-m303, CCR2-64I, and SDF1-3'A) in the Bahraini population. AIDS Res Hum Retroviruses 2009;25:973-7. 
|
| 15. | Gonzalez E, Dhanda R, Bamshad M, Mummidi S, Geevarghese R, Catano G, et al. Global survey of genetic variation in CCR5, RANTES, and MIP-1alpha: Impact on the epidemiology of the HIV-1 pandemic. Proc Natl Acad Sci U S A 2001;98:5199-204. 
|
| 16. | Su B, Sun G, Lu D, Xiao J, Hu F, Chakraborty R, et al. Distribution of three HIV-1 resistance-conferring polymorphisms (SDF1-3'A, CCR2-641, and CCR5-delta32) in global populations. Eur J Hum Genet 2000;8:975-9. 
|
| 17. | Smith MW, Dean M, Carrington M, Winkler C, Huttley GA, Lomb DA, et al. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science 1997;277:959-65. 
|
| 18. | Gonzalez E, Bamshad M, Sato N, Mummidi S, Dhanda R, Catano G, et al. Race-specific HIV-1 disease-modifying effects associated with CCR5 haplotypes. Proc Natl Acad Sci U S A 1999;96:12004-9. 
|
| 19. | Mummidi S, Ahuja SS, Gonzalez E, Anderson SA, Santiago EN, Stephan KT, et al. Genealogy of the CCR5 locus and chemokine system gene variants associated with altered rates of HIV-1 disease progression. Nat Med 1998;4:786-93. 
|
| 20. | An P, Martin MP, Nelson GW, Carrington M, Smith MW, Gong K, et al. Influence of CCR5 promoter haplotypes on AIDS progression in African-Americans. AIDS 2000;14:2117-22. 
|
| 21. | Callegari-Jacques SM, Grattapaglia D, Salzano FM, Salamoni SP, Crossetti SG, Ferreira ME, et al. Historical genetics: Spatiotemporal analysis of the formation of the Brazilian population. Am J Hum Biol 2003;15:824-34. 
|
| 22. | Pena SD. Reasons for banishing the concept of race from Brazilian medicine. Hist Cienc Saude Manguinhos 2005;12:321-46. 
|
| 23. | Munerato P, Azevedo ML, Sucupira MC, Pardini R, Pinto GH, Catroxo M, et al. Frequency of polymorphisms of genes coding for HIV-1 co-receptors CCR5 and CCR2 in a Brazilian population. Braz J Infect Dis 2003;7:236-40. 
|
| 24. | Vargas AE, Marrero AR, Salzano FM, Bortolini MC, Chies JA. Frequency of CCR5delta32 in Brazilian populations. Braz J Med Biol Res 2006;39:321-5. 
|
| 25. | Reiche EM, Ehara Watanabe MA, Bonametti AM, Morimoto HK, Akira Morimoto A, Wiechmann SL, et al. Frequency of CCR5-Delta32 deletion in human immunodeficiency virus type 1 (HIV-1) in healthy blood donors, HIV-1-exposed seronegative and HIV-1-seropositive individuals of southern Brazilian population. Int J Mol Med 2008;22:669-75. 
|
| 26. | Rigato PO, Hong MA, Casseb J, Ueda M, de Castro I, Benard G, et al. Better CD4+ T cell recovery in Brazilian HIV-infected individuals under HAART due to cumulative carriage of SDF-1-3'A, CCR2-V64I, CCR5-D32 and CCR5-promoter 59029A/G polymorphisms. Curr HIV Res 2008;6:466-73. 
|
| 27. | Majumder PP. People of India: Biological diversity and affinities. Evol Anthropo 1998;6:100-10. 
|
| 28. | Baig MM, Khan AA, Kulkarni KM. Mitochondrial DNA diversity in tribal and caste groups of Maharashtra (India) and its implication on their genetic origins. Ann Hum Genet 2004;68:453-60. 
|
| 29. | Majumder PP. Indian caste origins: Genomic insights and future outlook. Genome Res 2001;11:931-2. 
|
| 30. | Karve I. Hindu Society-An Interpretation. Poona: Deshmukh Prakashan; 1961. 
|
| 31. | Kasambi DD. The Culture and Civilization of Ancient India in Historical Outline. New Delhi: Vikas; 1964. 
|
| 32. | Roychoudhury S, Roy S, Basu A, Banerjee R, Vishwanathan H, Usha Rani MV, et al. Genomic structures and population histories of linguistically distinct tribal groups of India. Hum Genet 2001;109:339-50. 
|
| 33. | Singh S, Bhanu B. Anthropological Survey of India. Maharashtra: Popular Prakashan; 2004. p. 65. 
|
| 34. | Zhou H, Hickford JG, Fang Q. A two-step procedure for extracting genomic DNA from dried blood spots on filter paper for polymerase chain reaction amplification. Anal Biochem 2006;354:159-61. 
|
| 35. | Bernard NF, Yannakis CM, Lee JS, Tsoukas CM. Human immunodeficiency virus (HIV)-specific cytotoxic T lymphocyte activity in HIV-exposed seronegative persons. J Infect Dis 1999;179:538-47. 
|
| 36. | Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature 1996;381:661-6. 
|
| 37. | Dragic T, Trkola A, Lin SW, Nagashima KA, Kajumo F, Zhao L, et al. Amino-terminal substitutions in the CCR5 coreceptor impair gp120 binding and human immunodeficiency virus type 1 entry. J Virol 1998;72:279-85. 
|
| 38. | Eugen-Olsen J, Iversen AK, Garred P, Koppelhus U, Pedersen C, Benfield TL, et al. Heterozygosity for a deletion in the CKR-5 gene leads to prolonged AIDS-free survival and slower CD4 T-cell decline in a cohort of HIV-seropositive individuals. AIDS 1997;11:305-10. 
|
| 39. | Kozhekbaeva GM, Borodina TA, Borinskaia SA, Gusar VA, Feshchenko SP, Akhmetova VL, et al. Distribution of the HIV-1 resistance-conferring alleles (CCR5delta32, CCR2-64I, and SDF1 3'A) in Russian, Ukrainian, and Belarusian populations. Genetika 2004;40:1394-401. 
|
| 40. | Irtiza S, Dil-Afroze, Naykoo NA, Lateef C, Iqbal Q, Inayat SF, et al. Polymorphism in the CC-chemokine receptor-5 (CCR5) gene and risk of AIDS among Kashmiri population. J AIDS HIV Res 2011;3:103-6. 
|
| 41. | Apostolakis S, Baritaki S, Krambovitis E, Spandidos DA. Distribution of HIV/AIDS protective SDF1, CCR5 and CCR2 gene variants within Cretan population. J Clin Virol 2005;34:310-4. 
|
| 42. | Libert F, Cochaux P, Beckman G, Samson M, Aksenova M, Cao A, et al. The deltaccr5 mutation conferring protection against HIV-1 in Caucasian populations has a single and recent origin in Northeastern Europe. Hum Mol Genet 1998;7:399-406. 
|
| 43. | Leboute AP, de Carvalho MW, Simões AL. Absence of the deltaccr5 mutation in indigenous populations of the Brazilian Amazon. Hum Genet 1999;105:442-3. 
|
| 44. | Salem AH, Batzer MA. Distribution of the HIV resistance CCR5-Delta32 allele among Egyptians and Syrians. Mutat Res 2007;616:175-80. 
|
[Figure 1]
[Table 1]
|






