|
 
 |
|
CASE REPORT |
|
|
|
| Year : 2013 | Volume
: 19
| Issue : 1 | Page : 96-100 |
| |
A novel deletion cluster at 13q14.2-q21.33 in an 80-year man with late onset leukemia: Clinical and molecular findings
Kiran Kumar1, Sujit Maiti1, Christina A Castellani1, Richard O'Reilly2, Shiva M Singh3
1 Department of Biology, Molecular Genetics Unit, The University of Western Ontario, London, ON, Canada
2 Department of Psychiatry and London Health Sciences Center, London, ON, Canada
3 Department of Biology, Molecular Genetics Unit, The University of Western Ontario; Department of Psychiatry and London Health Sciences Center, London, ON, Canada
| Date of Web Publication | 4-Jun-2013 |
Correspondence Address:
Shiva M Singh
Department of Biology, Molecular Genetics Unit, The University of Western Ontario, Richmond Street, London, ON, N6A 5B7
Canada
 Source of Support: Canadian Institute of Health Research and Schizophrenia Society of Ontario., Conflict of Interest: None
DOI: 10.4103/0971-6866.112916

 Abstract Abstract | | |
Chromosomal deletions are among the most common genetic events observed in hematologic malignancies; loss of genetic material is regarded as a hallmark of putative tumor suppressor gene localization. We have identified an unusual cluster of deletions at 13q14.2-13q21.33 in an 80-year-old father of a monozygotic twin pair discordant for schizophrenia, who developed chronic leukemia (CLL) at age 69.
Materials and Methods: The breakpoints for individual deletions in this cluster was identified by Affymetrix Human Array 6.0 screening.
Results: The deleted segments harbours a number of genes, most associated with cancer as well as a high concentration of LINEs, SINEs and related repeats. The derived chromosome represents an intra-chromosomal re-arrangement that quickly overtook blood progenitor cells probably before age 69 as a cause of CLL.
Conclusions: The study highlights the role of ongoing de novo changes at susceptible sites, such as repeat rich regions, in the human genome. Also, it argues for the involvement of genes/deletions in the 13q(14.2-21.33) region in the development of CCL.
Keywords: Copy number variations, deletions, de novo mutations, LINES, leukemia, SINES
How to cite this article:
Kumar K, Maiti S, Castellani CA, O'Reilly R, Singh SM. A novel deletion cluster at 13q14.2-q21.33 in an 80-year man with late onset leukemia: Clinical and molecular findings. Indian J Hum Genet 2013;19:96-100 |
How to cite this URL:
Kumar K, Maiti S, Castellani CA, O'Reilly R, Singh SM. A novel deletion cluster at 13q14.2-q21.33 in an 80-year man with late onset leukemia: Clinical and molecular findings. Indian J Hum Genet [serial online] 2013 [cited 2016 May 24];19:96-100. Available from: http://www.ijhg.com/text.asp?2013/19/1/96/112916 |
 Introduction Introduction | |  |
Genome-wide, high-resolution arrays are rapidly becoming a reliable method of molecular investigation across individuals, families, and populations. Developed just a few years ago, they have now become a primary method for assessment of the quantitative and qualitative genomic variations involving single nucleotide polymorphisms, copy number variations (CNVs), and insertions and deletions. They have been instrumental in establishing genome-wide associations in a variety of disorders including cancers, and congenital anomalies. Further, the results have helped identify the involvement of specific genes and genomic alterations in a number of cancers. This case report represents one such example and deals with identification of a major genomic rearrangement by Affymetrix Human Array 6.0 in an 80-year-old male who developed leukemia at 69 years of age.
The subject was identified and recruited for this study as the father of a monozygotic twin pair discordant for schizophrenia and clinically assessed by Dr. Richard O'Reilly (Psychiatrist). He gave informed consent and provided blood and buccal cells to be used in this research protocol that has been approved by the Committee on Research Involving Human Subjects at the University of Western Ontario.
 Case Report Case Report | |  |
The subject was an 80-year-old at the time of assessment. He had a high school diploma and had worked in sales until he retired. The subject had no major illness until he was diagnosed with leukemia at age 69. At age 70 he was noted to be hypertensive and 3 years later had a quadruple coronary bypass. Immediately following his coronary artery bypass the subject developed low mood and anxiety. And then after discharge from hospital he started to experience panic attacks and was prescribed lorazepam for 2 months. At age 80 the subject's gallbladder ruptured and he developed septicemia. He had emergency surgery following which he lost weight, became debilitated and experienced further symptoms of depression. The subject received the last course of nine chemotherapy treatments for leukemia 6 months before the blood was drawn for genetic assessment. At the time when the blood was drawn the subject was taking atorvastin sodium daily to treat hypercholesterolemia.
 Results and Discussion Results and Discussion | |  |
The subject's leucocyte genomic DNA was hybridized to the Affymetrix Human Array 6.0 following the manufacturer's protocol at the London Regional Genomics Center, The University of Western Ontario. Briefly, 5 μg of genomic DNA was labeled and hybridized to the array. Calls for CNVs were made using the Affymetrix Genotyping Console 4.0 as well as Partek H Genotyping Suite™ software suites. In both cases, the CNVs were identified by continuity of markers on a segment. Two CNVs that overlapped by 50% in the two methods of data analysis were given the same identity. Every measure was undertaken to avoid inclusion of false positives including correction for segmental duplications. The CNVs identified were further assessed by comparison to the Database of Genomic Variants ( http://projects.tcag.ca/variation/ ) and annotated with gene symbols by importing the annotation file from the UCSC genome browser (NCBI36/hg 18). This analysis identified an unusual cluster of gains and losses on 13q that forms the focus of this report.
[Table 1] shows the cytogenetic and molecular (nt) breakpoints as well as the size of deleted fragments and gene affected. The genomic features of this abnormality as shown in [Figure 1], suggests that it harbors a number of genes, some labeled in this figure. More important the genes affected are implicated in a number of pathways [Table 2]. It is apparent from this table that a large number of genes are known to play a role in cancers, cell cycle regulation. Further, the region harbors several MicroRNA coding genes, which function in cell survival, proliferation, differentiation, and angiogenesis and is the primary target of genomic amplification that occurs in several lymphomas and solid tumors. [1] A recent study by Parker et al. [2] has assessed 13q deletions in chronic lymphocytic leukemia (CLL) using genomic profiling. They identified 205 copy number alterations on chromosome 13 in 132 cases. These deletions were highly heterogeneous (845 Kb to 96.2 Mb) in size. They also identified two breakpoint cluster regions within short interspersed nuclear elements proximal to DLEU5 (TRIM13) and within long interspersed nuclear elements/L1 repeats distal to GUCY1B2.
Forty breakpoints, almost all associated with the distribution of LINES and SINES [Figure 1]. Parker et al. also suggested that the larger deletions (Class II), as seen in our subject have an increased risk of disease progression (odds ration = 12.3; P = 0.005). Interestingly, deletions on the long arm of chromosome-13 have also been reported with mental and motor retardation, craniofacial dysmorphic facial appearance and various congenital malformations. [3],[4],[5],[6] Furthermore, del13q is known to manifest with a range of abnormalities, including, retinoblastoma, mental and growth retardation, brain malformations, heart defects, distal limb deformities, and digestive, and urogenital abnormalities ( www.diseasesdatabase.com ). Interestingly, none of these symptoms are apparent in this case.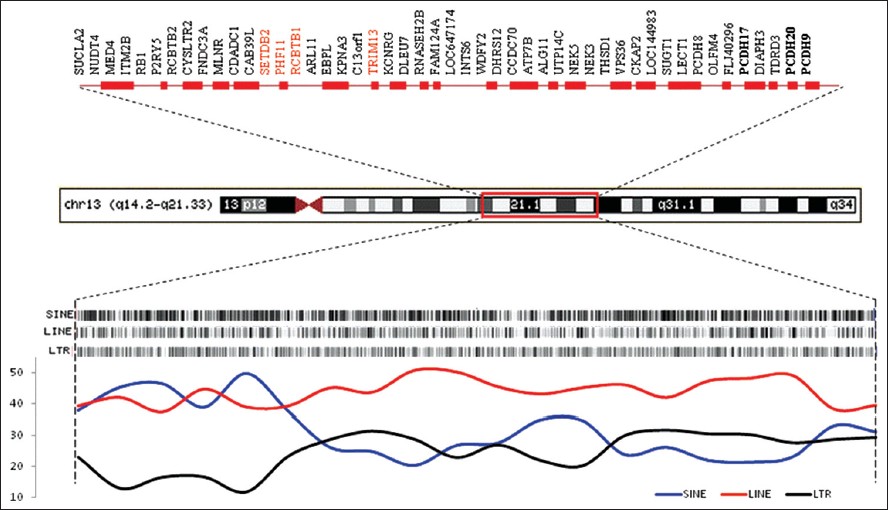 | Figure 1: Ideogram of the chromosome 13q deletion (14.2 - 21.33) depicting the annotated refseq genes accessed from map viewer (top) and distribution of major class of repeats in the region (bottom). Genes listed in red are reported in schizophrenia database
Click here to view |
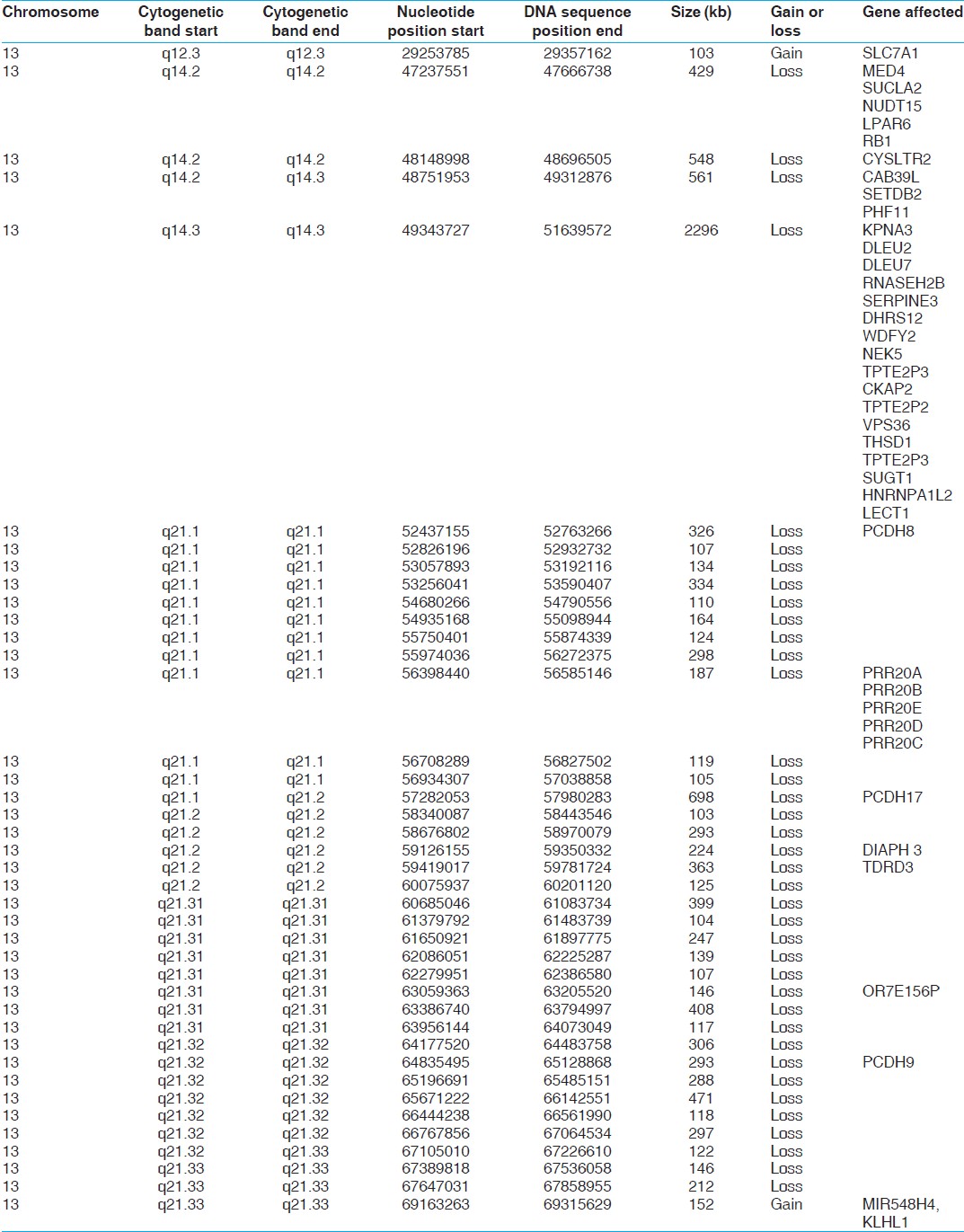 | Table 1: Genomic (cytogenetic and nucleotide numbers) breakpoints, corresponding size of deletions and genes affected in the 13q (14.2 - 21.33) region in the subject
Click here to view |
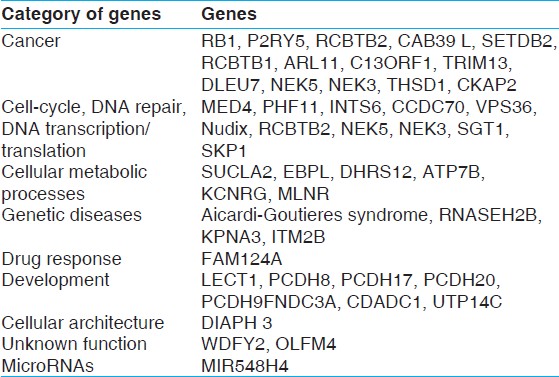 | Table 2: Functional categories of genes deleted at the 13q (14.2 - 21.33) region. They predominately include genes involved in cancers, cell cycle and development. Furthermore, a number of genes listed are known to be involved in CCL
Click here to view |
Chromosomal deletions are among the most common genetic events observed in hematologic malignancies. It includes differential loss of genetic material from 13q in lymphoid neoplasias, in non-Hodgkin's lymphoma and in chronic lymphoproliferative diseases. [7] The presence the complex deletion involving 13q14.2-13q21.33 in our subject is best explained by the origin of this abnormality later in life, probably before age 69 when he was diagnosed with CLL. Given the genes involved, it is argued that this abnormality must have provided proliferative advantage that led to the development of CLL at age 69. Given that, the observed deletion is large and contains ~50 genes including genes implicated in CLL, this feature may have accounted for the fast progression of CLL in this patient. Such a speculation is compatible with the concentration and specific location of the LINE1, SINE, and related repeats [Figure 1] in 13q14.2-21.33. This feature will make this genomic region prone to intra-chromosomal recombination during mitotic DNA replication. This would support recent reports [8] that de novo mutations that are operational during the life-time [9] can occur both randomly and in response to external challenges. The phenotypic description of deletion 13q syndrome is dependent on the location and size of the deleted segment. At present, the syndrome is divided into three groups based on the deletion's location relative to chromosomal band 13q32. Groups 1 (proximal to q32) and 2 (including q32) have shown distinctive phenotypes including mental retardation and growth deficiency. [10] However, del13q manifests with a range of abnormalities, including, retinoblastoma, mental and growth retardation, brain malformations, heart defects, distal limb deformities, and digestive, and urogenital abnormalities have been reported previously (www.diseasesdatabase.com). This case adds yet another example of large deletions on 13q that account for the initiation and rapid progression of chronic lymphocytic leukemia.
 Conclusion Conclusion | |  |
The human genome continues to undergo genomic changes randomly and in response to a variety of internal and external factors. We report a cluster of somatic deletions covering 13q14.2-13q21.33, along with individual breakpoints and genes affected in a male that the most likely has caused progressing CLL. Such results support the dynamic nature of the human genome and the role of specific genomic aberrations in leukemia.
 References References | |  |
| 1. | Olive V, Jiang I, He L. mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol 2010;42:1348-54. 
|
| 2. | Parker H, Rose-Zerilli MJ, Parker A, et al.: 13q deletion anatomy and diseaseprogression in patients with chronic lymphocytic leukemia. Leukemia 2011Mar; 25(3):489-97. 
|
| 3. | Balci S, Yuksel Konuk B, Atik F, Oguz AK, Ergun MA, Baltaci V, et al. Partial deletion of the long arm of chromosome 13 (q32q33.2) associated with mental retardation, choanal atresia and fish mouth. Genet Couns 2010;21:317-24. 
|
| 4. | Quélin C, Bendavid C, Dubourg C, de la Rochebrochard C, Lucas J, Henry C, et al. Twelve new patients with 13q deletion syndrome: Genotype-phenotype analyses in progress. Eur J Med Genet 2009;52:41-6. 
|
| 5. | Roche A, Mora J, Perez Mdel M, Gean E, Perez B, O'Callaghan M, et al. Axenfeld-Rieger ocular anomaly and retinoblastoma caused by constitutional chromosome 13q deletion. Pediatr Blood Cancer 2010;54:480-2. 
|
| 6. | Takahashi K, Oka A, Mizuguchi M, Saitoh M, Takita J, Sato A, et al. Interstitial deletion of 13q14.13-q32.3 presenting with Arima syndrome and bilateral retinoblastoma. Brain Dev 2011;33:353-6. 
|
| 7. | Mitelman F, Mertens F, Johansson B. A breakpoint map of recurrent chromosomal rearrangements in human neoplasia. Nat Genet 1997;15:417-74. 
|
| 8. | Maiti S, Kumar KH, Castellani CA, O'Reilly R, Singh SM. Ontogenetic de novo copy number variations (CNVs) as a source of genetic individuality: Studies on two families with MZD twins for schizophrenia. PLoS One 2011;6:e17125. 
|
| 9. | Laski JR. New mutations and intellectual function. Nat Genet 2010;42:1036-8. 
|
| 10. | Lance EI, DuPont BR, Holden KR. Expansion of the deletion 13q syndrome phenotype: A case report. J Child Neurol 2007;22:1124-7. 
|
[Figure 1]
[Table 1], [Table 2]
|






