|
 
 |
|
ORIGINAL ARTICLE |
|
|
|
| Year : 2013 | Volume
: 19
| Issue : 2 | Page : 165-170 |
| |
Multiplex ligation-dependant probe amplification study of children with idiopathic mental retardation in South India
Neetha John1, Moka Rajasekhar1, Katta Mohan Girisha2, Podila Satya Venkata Narasimha Sharma3, Puthiya Mundyat Gopinath1
1 Division of Biotechnology, Manipal Life Sciences Centre, Manipal University, Manipal, Karnataka, India
2 Department of Pediatrics, Kasturba Medical College, Manipal University, Manipal, Karnataka, India
3 Department of Psychiatry, Kasturba Medical College, Manipal University, Manipal, Karnataka, India
| Date of Web Publication | 5-Aug-2013 |
Correspondence Address:
Moka Rajasekhar
Division of Biotechnology, Manipal Life Sciences Centre, Manipal University, Manipal - 576 104, Karnataka
India
 Source of Support: Indian Council Medical Research (ICMR), New Delhi, Conflict of Interest: None
DOI: 10.4103/0971-6866.116115

 Abstract Abstract | | |
Background: Mental retardation (MR) is a heterogeneous dysfunction of the central nervous system exhibiting complex phenotypes and has an estimated prevalence of 1-3% in the general population. However, in about 50% of the children diagnosed with any form of intellectual disability or developmental delay the cause goes undetected contributing to idiopathic intellectual disability.
Materials and Methods: A total of 122 children with developmental delay/MR were studied to identify the microscopic and submicroscopic chromosome rearrangements by using the conventional cytogenetics and multiplex ligation dependent probe amplification (MLPA) analysis using SALSA MLPA kits from Microbiology Research Centre Holland [MRC] Holland.
Results: All the recruited children were selected for this study, after thorough clinical assessment and metaphases prepared were analyzed by using automated karyotyping system. None was found to have chromosomal abnormality; MLPA analysis was carried out in all subjects and identified in 11 (9%) patients.
Conclusion: Karyotype analysis in combination with MLPA assays for submicroscopic micro-deletions may be recommended for children with idiopathic MR.
Keywords: Developmental delay, mental retardation, micro-deletions, multiplex ligation dependent probe amplification, SH3 and multiple ankyrin repeat domains 3
How to cite this article:
John N, Rajasekhar M, Girisha KM, Sharma PV, Gopinath PM. Multiplex ligation-dependant probe amplification study of children with idiopathic mental retardation in South India. Indian J Hum Genet 2013;19:165-70 |
How to cite this URL:
John N, Rajasekhar M, Girisha KM, Sharma PV, Gopinath PM. Multiplex ligation-dependant probe amplification study of children with idiopathic mental retardation in South India. Indian J Hum Genet [serial online] 2013 [cited 2016 May 24];19:165-70. Available from: http://www.ijhg.com/text.asp?2013/19/2/165/116115 |
 Introduction Introduction | |  |
Intellectual disability, also called as mental retardation (MR) is a generalized disorder branded by significantly impaired cognitive functioning existing concurrently with related limitations in two or more of the adaptive skill areas such as community use, self-direction, health and safety, functional academics, leisure, and work and appears before the age of 18 years. With regard to the intellectual criteria for the diagnosis of MR, intelligence is generally defined by an intelligent quotient (IQ) test score of approximately 70 or below. Levels of MR has been classified by Diagnostic and Statistical Manual of Mental Disorders IV-Text Revision (DSM-IV-TR) based on IQ scores; mild to moderate MR being >50 and moderate to severe MR is <50. The prevalence of MR is 2-3% of the general population. [1] MR may be caused by a wide range of factors that, together, contribute to its pathogenesis. In different studies on the etiology, the diagnostic yield appears to be highly variable. [2] This variation is likely attributable to difference in methodology, classification and terms used for diagnosis. [3] Genetic disorders most commonly found in individuals with developmental delay/MR are chromosomal aberrations at about 10%. [4],[5] Down syndrome (DS) is the most common with MR associated chromosomal abnormality, [6] Fragile X syndrome (FXS) is 2 nd most common form and inherited MR. Besides DS and FXS, chromosome aberrations are so common. Conventional cytogenetics is the primary diagnostic tool to detect chromosomal abnormalities. However, with the advent of novel genetic techniques, submicroscopic, and subtelomeric chromosomal rearrangements were detected. The present investigation was aimed at studying submicroscopic chromosomal variations in patients with idiopathic MR by using multiplex ligation dependent probe amplification (MLPA) and also compared our results with previous studies [Table 1] in subjects with MR.
 Materials and Methods Materials and Methods | |  |
A total of 122 subject of age group between 3 years and 18 years with developmental disabilities were recruited. The IQ of all the study subjects (above 5 years) were determined to be below 70 (Wechsler intelligence scale for children). The etiology of developmental delay/or intellectual disability was found out using inclusion and exclusion criteria [7] such as (1) dysmorphologic examination (e.g., minor anomalies and malformations), (2) neurologic examination (e.g., electroencephalography, neuroimaging), (3) cytogenetic screening (G-bands by Trypsin using Giemsa - GTG banded metaphases) and (4) fragile X screening (FraX site using M199 medium and polymerase chain reaction (PCR) analysis for Fragile X Mental Retardation 1 (FMR1) gene, Cytosine-Guanine-Guanine (CGG triplet repeats). Cytogenetic analysis was carried out as per standardized method [8] in every recruited patient. Genomic DNA was extracted and purified from peripheral blood samples by using standardized phenol-chloroform method as well as Qiagen DNA extraction kit. A total of 10 normal controls (5 male and 5 female) were included in the study as per the criteria of SALSA MLPA kits viz. P036, P070, P245 A2 and MR X106B1. The SALSA P036 and P070 probe mixture contains 1 MLPA probe for each submicroscopic subtelomeric region. The SALSA P245 A2 contains probes targeting regions specific for intellectual disability and MR X 106 B1 contains probes, which can detect subtle rearrangements on X chromosome related to X Linked Mental Retardation (XLMR).
In the study group, MLPA was used to assess the presence of cryptic submicroscopic chromosomal imbalances in 122 patients with unexplained developmental delay/intellectual disability. A set of SALSA MLPA - P036, P070, P245 A2, and MR X 106B1 test kits (MRC-Holland, Amsterdam, The Netherlands; www.mrc-holland.com ) were employed. The regions hence studied are shown in [Table 2]. The MLPA analysis was carried out as described by Schouten et al., [9] with minor modifications. Reaction products were electrophoresed in the ABI-PRISM 3130 genetic analyzer (Applied Biosystems, USA). For reference, the normal DNAs were pooled into two mixtures (one for each sex) to avoid individual variations and obtain average normal patterns. These standard patterns, one each for male and female were subsequently employed to perform the data analysis in all the studied samples. The signal intensities of the PCR products were determined by using GeneMapper v4.0 software (Applied Biosystems). A spreadsheet was developed in Microsoft Excel to process the data for the control samples (with no family history of intellectual disability) efficiently. The raw data for the 10 control samples were processed by dividing the signal intensity of each probe by the averaged signal intensity of all peaks in each sample. Later, each peak was normalized by dividing the obtained average peak by the corresponding average peak from the pooled control samples for which GeneMarker (Soft-genetics, Pennsylvania) was employed. The values are expected to be around 1.0 for sequences as the control samples are expected to have a normal dosage.
The variability of the normalized values for each probe in these control samples were determined and were plotted using Origin as shown in [Figure 1] and the probes showed a homogenous behavior, and the normalized values clustered around one although slight differences on skewness and range were observed for some of them. Hence, not all probes conferred the same reliability taking into account the borderline results that were obtained. The results obtained with the same assay in normal men and women confirmed the reliability of our data. The study was approved by the Institutional Ethical Committee and a written informed consent was obtained from all participants.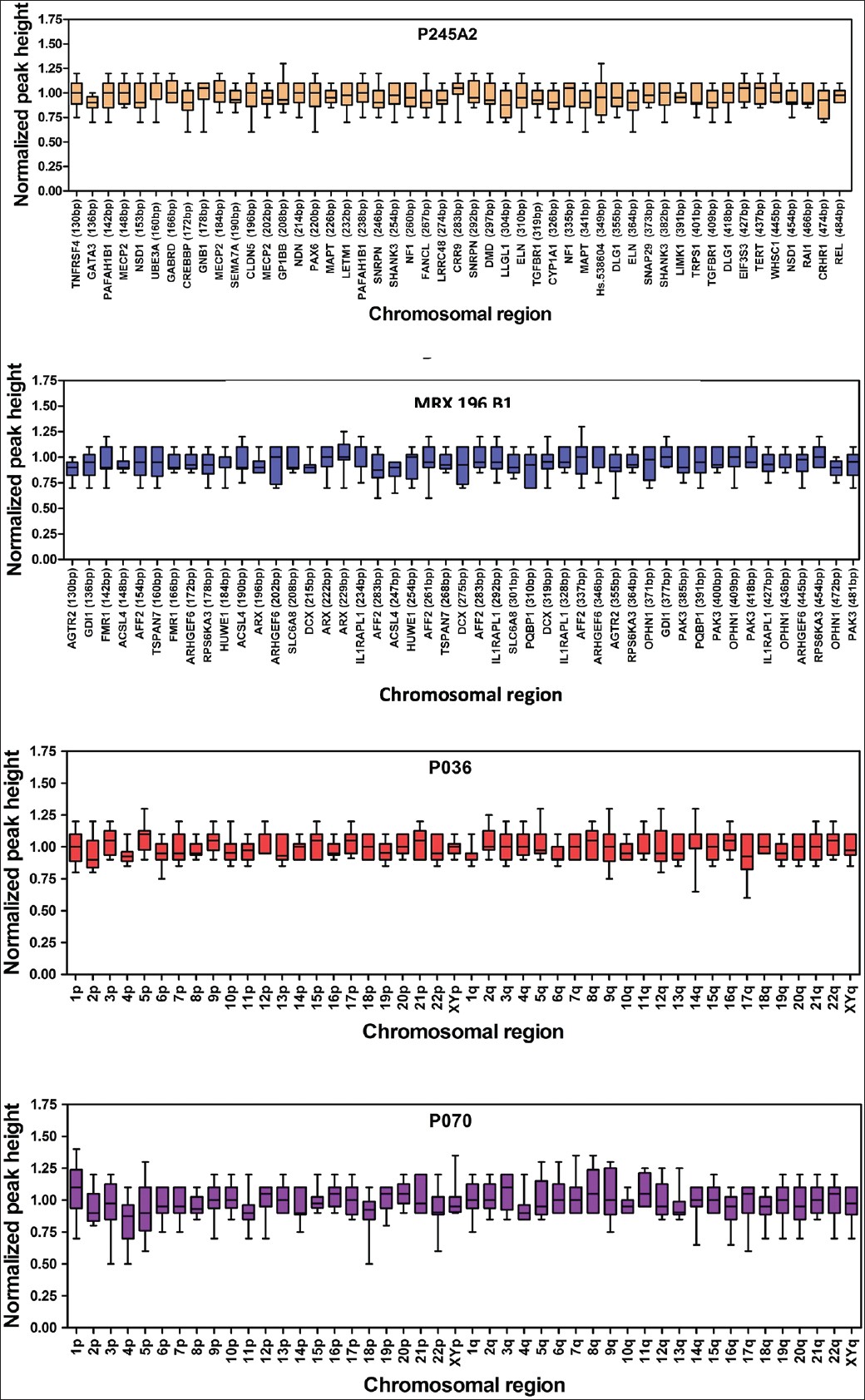 | Figure 1: Box plots of the normalized values for each probe from P245A2, MRX106B1, P070 and P036 in the control samples
Click here to view |
 Results Results | |  |
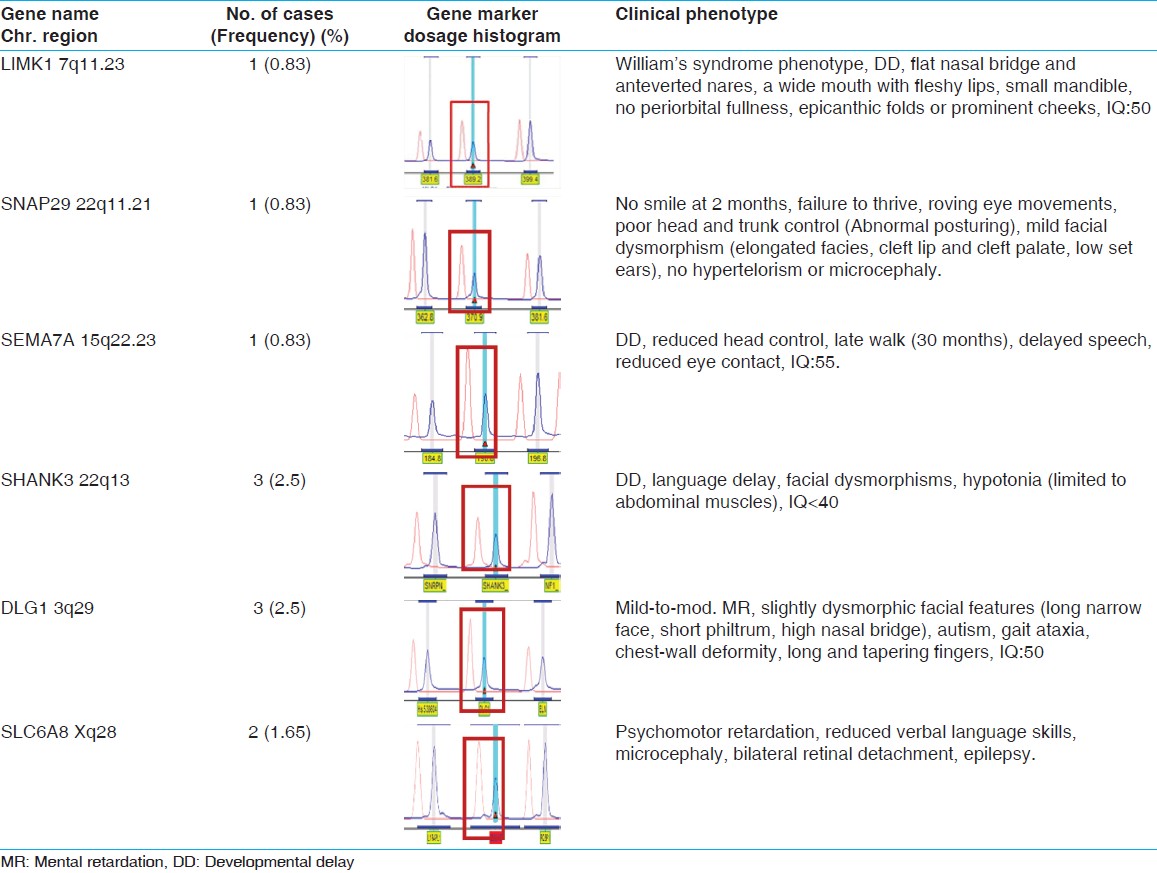
A total of 122 children with developmental delay/or MR were selected to study cytogenetic and MLPA analysis [Figure 2] and [Table 3] after a thorough clinical investigation. All patients had apparently normal karyotypes and showed no rearrangements. SALSA MLPA P245-A2 kit was applied to 122 patients and P 070 and P 036-75 patients each. MRX106-B1 kit was used on 60 cases with suspected phenotype for X-linked MR. [Table 4] shows a summary of all detected micro-deletions. Three patients showed micro-deletion in SHANK3 using P245-A2 kit, which has a clear clinical significance as it overlaps with a known micro-deletion syndrome. However, micro-deletions of DLG1 detected in three patients, LIMK1, SNAP29 and SEMA7A detected in one patient each, which were reported by two study groups. [10],[11] No subtelomeric deletions or duplication were observed in the present study group.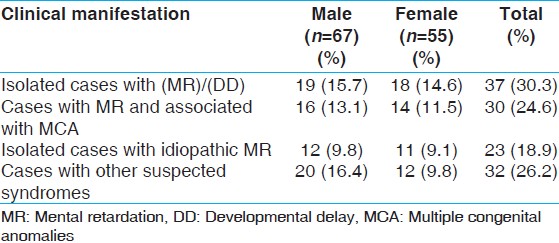 | Table 3: Clinical manifestation of the samples (n=122) collected and their frequencies
Click here to view |
 | Table 4: Details of clinical phenotypes and MLPA analysis of eleven patients (9%)
Click here to view |
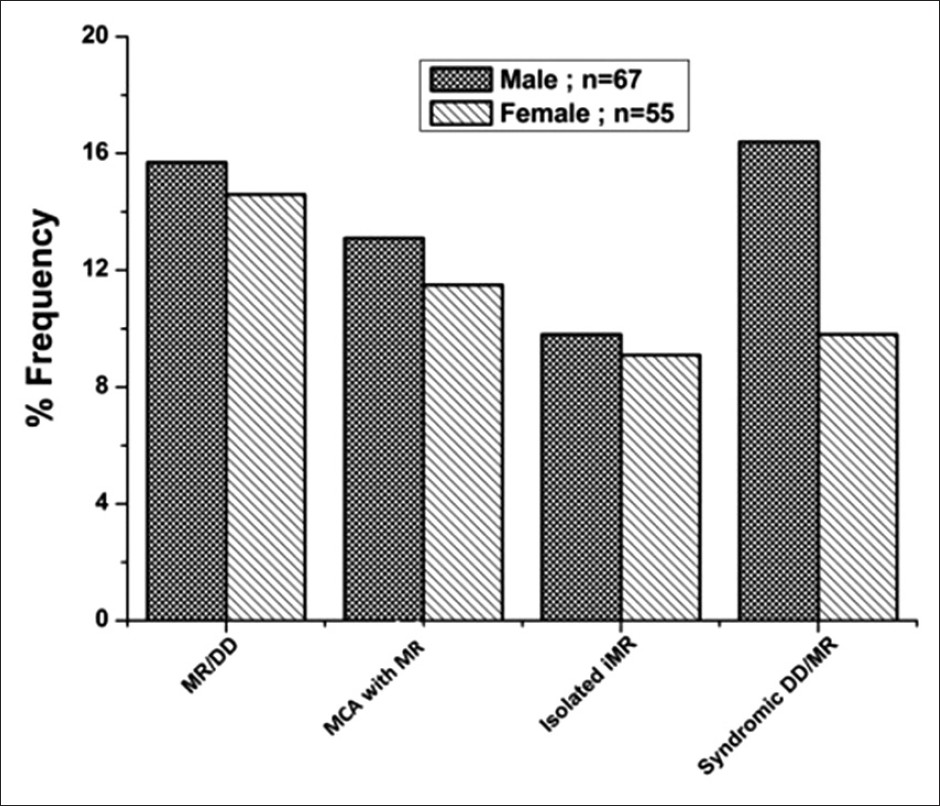 | Figure 2: Frequency distribution of study patients with mild to moderate intellectual disability
Click here to view |
 Discussion Discussion | |  |
In this study, we present the results of the submicroscopic screening by MLPA in 122 unexplained intellectual disability patients with normal Karyotypes. The results of the present study highlight the benefit of submicroscopic genomic screening in unexplained MR. The frequency of abnormalities in this study is about 9% of the patients and is comparable to previous studies. [12],[13] We identified eleven submicroscopic deletions in this study as shown in [Table 4]. However, parents were not available for inheritance studies for these subjects and del (7q11.23) was identified in one patient who was suspected to have Williams syndrome. Two genes in particular, ELN and LIMK1, been reported to be important in causing some of the characteristic symptoms of Williams syndrome. However, in this patient LIMK1 gene may be associated with mild MR. [14] 22q11 deletion syndrome is one of the most common cardiac abnormalities and rarely causes learning disabilities and mild MR, [15],[16] however, the presented patient had mild facial dysmorphism, failure to thrive, no evidence of hypertelorism and microcephaly who diagnosed as DiGeorge syndrome. In another group, we identified a micro-deletion at subtelomeric region of 22q13 in 3 children with developmental delay including language development in two of them and facial dysmorphism. SHANK3 is one of the genes disrupted in patients with the 22q13.3 deletion syndrome. The deletion syndrome is characterized by neonatal hypotonia, global developmental delay, normal to accelerated growth, absent to severely delayed speech, autistic behavior, and minor dysmorphic features. [17]
A 3½-year-old boy with developmental delay reported to be del (15q24.1), which usually associated with two genes main genes of SEMA7A and CPLX3 that may play an important role in brain development and may contribute to the cognitive disability in individuals with this deletion. [10] Similarly, del (3q29) was identified in three patients with mild to moderate intellectual disability and autistic phenotypes. This deletion mainly encompasses 22 genes, including PAK2 and DLG1, and they are also known to play a critical role in MR and related phenotypes. [11] Detection rate for submicroscopic imbalance can be assessed by MLPA, although none of the subtelomere probes yielded positive results, only the abnormalities found by micro-deletion/XLMR testing could be definitively established as of clinical significance. We also identified Xq28 deletion in a male patient with psychomotor retardation by using MRX 106B1 kit.
This paper reports a group of patients with developmental delay and/or mild to moderate intellectual disability recruited to find out genetic causes using cytogenetic and MLPA analysis. The purpose is to unearth the submicroscopic subtelomere deletions that could be missed by routine conventional cytogenetics. Recent methodologies for assessing the genomic imbalance at submicroscopic subtelomeres concluded that MLPA to be a robust technique in a diagnostic set-up although the use of real-time PCR and microarrays may become more widespread with their availability on an affordable commercial platform. [18],[19] The strategy of replacing conventional cytogenetic analysis by MLPA in a diagnostic set-up has been suggested in a recent study, if follow-up by microarray analysis is feasible as it may prove effective in terms of detection rate and cost-effectiveness. [20] MLPA is an extremely efficient, medium-throughput technique. For example, the cytogenetic findings in this study showed normal karyotypes in 99% of cases but with MLPA, the study detected in 9% of the patients with micro-deletions as given in [Table 4] Hence, MLPA method gives a better yield in comparison with karyotype analysis. MLPA can also be considered as an ideal approach until microarray testing gets validated as routine diagnostic tool, and becomes economical enough to replace karyotyping as the 1 st test for idiopathic MR patients.
 References References | |  |
| 1. | Moeschler JB, Shevell M, American academy of pediatrics committee on genetics. Clinical genetic evaluation of the child with mental retardation or developmental delays. Pediatrics 2006;117:2304-16. 
|
| 2. | Curry CJ, Stevenson RE, Aughton D, Byrne J, Carey JC, Cassidy S, et al. Evaluation of mental retardation: Recommendations of a consensus conference: American College of Medical Genetics. Am J Med Genet 1997;72:468-77. 
|
| 3. | Moog U. The outcome of diagnostic studies on the etiology of mental retardation: Considerations on the classification of the causes. Am J Med Genet A 2005;137:228-31. 
|
| 4. | Flint J, Wilkie AO. The genetics of mental retardation. Br Med Bull 1996;52:453-64. 
|
| 5. | van Karnebeek CD, Jansweijer MC, Leenders AG, Offringa M, Hennekam RC. Diagnostic investigations in individuals with mental retardation: A systematic literature review of their usefulness. Eur J Hum Genet 2005;13:6-25. 
|
| 6. | Roizen NJ, Patterson D. Down's syndrome. Lancet 2003;361:1281-9. 
|
| 7. | de Vries BB, White SM, Knight SJ, Regan R, Homfray T, Young ID, et al. Clinical studies on submicroscopic subtelomeric rearrangements: A checklist. J Med Genet 2001;38:145-50. 
|
| 8. | Moorhead PS, Nowell PC, Mellman WJ, Battips DM, Hungerford DA. Chromosome preparations of leukocytes cultured from human peripheral blood. Exp Cell Res 1960;20:613-6. 
|
| 9. | Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res 2002;30:e57. 
|
| 10. | El-Hattab AW, Zhang F, Maxim R, Christensen KM, Ward JC, Hines-Dowell S, et al. Deletion and duplication of 15q24: Molecular mechanisms and potential modification by additional copy number variants. Genet Med 2010;12:573-86. 
|
| 11. | Mulle JG, Dodd AF, McGrath JA, Wolyniec PS, Mitchell AA, Shetty AC, et al. Microdeletions of 3q29 confer high risk for schizophrenia. Am J Hum Genet 2010;87:229-36. 
|
| 12. | Rooms L, Reyniers E, van Luijk R, Scheers S, Wauters J, Ceulemans B, et al. Subtelomeric deletions detected in patients with idiopathic mental retardation using multiplex ligation-dependent probe amplification. Hum Mutat 2004;23:17-21. 
|
| 13. | Kriek M, Knijnenburg J, White SJ, Rosenberg C, den Dunnen JT, van Ommen GJ, et al. Diagnosis of genetic abnormalities in developmentally delayed patients: A new strategy combining MLPA and array-CGH. Am J Med Genet A 2007;143:610-4. 
|
| 14. | Bellugi U, Lichtenberger L, Mills D, Galaburda A, Korenberg JR. Bridging cognition, the brain and molecular genetics: Evidence from Williams syndrome. Trends Neurosci 1999;22:197-207. 
|
| 15. | Kobrynski LJ, Sullivan KE. Velocardiofacial syndrome, DiGeorge syndrome: The chromosome 22q11.2 deletion syndromes. Lancet 2007;370:1443-52. 
|
| 16. | Bales AM, Zaleski CA, McPherson EW. Newborn screening programs: Should 22q11 deletion syndrome be added? Genet Med 2010;12:135-44. 
|
| 17. | Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet 2007;39:25-7. 
|
| 18. | Galasso C, Lo-Castro A, El-Malhany N, Curatolo P. "Idiopathic" mental retardation and new chromosomal abnormalities. Ital J Pediatr 2010;36:17. 
|
| 19. | Stuppia L, Antonucci I, Palka G, Gatta V. Use of the MLPA assay in the molecular diagnosis of gene copy number alterations in human genetic diseases. Int J Mol Sci 2012;13:3245-76. 
|
| 20. | Hills A, Ahn JW, Donaghue C, Thomas H, Mann K, Ogilvie CM. MLPA for confirmation of array CGH results and determination of inheritance. Mol Cytogenet 2010;3:19. 
|
[Figure 1], [Figure 2]
[Table 1], [Table 2], [Table 3], [Table 4]
|






