|
 
 |
|
ORIGINAL ARTICLE |
|
|
|
| Year : 2013 | Volume
: 19
| Issue : 3 | Page : 311-314 |
| |
Phenotypic spectrum in uniparental disomy: Low incidence or lack of study?
Arpan D Bhatt1, Thomas Liehr2, Sonal R Bakshi1
1 Institute of Science, Nirma University, Ahmedabad, Gujarat, India
2 Jena University Hospital, Friedrich Schiller University, Institute of Human Genetics, 07740 Jena, Germany
| Date of Web Publication | 30-Oct-2013 |
Correspondence Address:
Sonal R Bakshi
Institute of Science, Nirma University, Sarkhej-Gandhinagar Highway, Chandlodia, Gota, Ahmedabad - 382 481, Gujarat
India
 Source of Support: Nirma Education and Research Foundation, Conflict of Interest: None
DOI: 10.4103/0971-6866.120819

 Abstract Abstract | | |
Context: Alterations in the human chromosomal complement are expressed phenotypically ranging from (i) normal, via (ii) frequent fetal loss in otherwise normal person, to (iii) sub-clinical to severe mental retardation and dysmorphism in live births. A subtle and microscopically undetectable chromosomal alteration is uniparental disomy (UPD), which is known to be associated with distinct birth defects as per the chromosome involved and parental origin. UPD can be evident due to imprinted genes and/or activation of recessive mutations.
Aims: The present study comprises of data mining of published UPD cases with a focus on associated phenotypes. The goal was to identify non-random and recurrent associations between UPD and various genetic conditions, which can possibly indicate the presence of new imprinted genes.
Settings and Design: Data mining was carried out using the homepage "http://www.fish.uniklinikum-jena.de/UPD.html," an online catalog of published cases with UPD.
Materials and Methods: The UPD cases having normal karyotype and with or without clinical findings were selected to analyze the associated phenotypes for each chromosome, maternal or paternal involved in UPD.
Results: Our results revealed many genetic conditions (other than the known UPD syndromes) to be associated with UPD. Even in cases of bad obstetric history as well as normal individuals chance detection of UPD has been reported.
Conclusions: The role of UPD in human genetic disorders needs to be studied by involving larger cohorts of individuals with birth defects as well as normal population. The genetic conditions were scrutinized in terms of inheritance patterns; majority of these were autosomal recessive indicating the role of UPD as an underlying mechanism.
Keywords: Autosomal recessive, birth defects, data mining, phenotypic expression, uniparental disomy
How to cite this article:
Bhatt AD, Liehr T, Bakshi SR. Phenotypic spectrum in uniparental disomy: Low incidence or lack of study?. Indian J Hum Genet 2013;19:311-4 |
How to cite this URL:
Bhatt AD, Liehr T, Bakshi SR. Phenotypic spectrum in uniparental disomy: Low incidence or lack of study?. Indian J Hum Genet [serial online] 2013 [cited 2016 May 24];19:311-4. Available from: http://www.ijhg.com/text.asp?2013/19/3/311/120819 |
 Introduction Introduction | |  |
The causative role of chromosomal anomalies in patients with inherited disorders is well-established. In addition to the known syndromes, there are other genetic conditions involving a large number of microscopic (structural and numerical karyotypic anomalies) as well as submicroscopic genetic alterations. The submicroscopic anomalies include micro deletions, point mutations, genomic imprinting and uniparental disomy (UPD). UPD can either be maternally or paternally derived. The concept of UPD was initially introduced by Eric Engel in 1980, [1] the experimental evidence was obtained when UPD(7)mat was demonstrated in a cystic fibrosis patient. [2] This was followed by more reports of UPD in other syndromes such as Angelman syndrome (AS), Prader Willi syndrome (PWS), Beckwith-Wiedemann syndrome More Details (BWS) or Silver Russel-syndrome (SRS) and were non-random enough to have diagnostic implications. [3]
Studying the role of UPD in a disease phenotype is complex due to various factors. First, it has been observed that even in well-defined syndromes [Table 1] the incidence of UPD along with a disease phenotype is 2-5% in AS, 25% in PWS, 20% in BWS, and 5% in SRS. [4] Second, based on the chromosome involved in paternal or maternal UPD, the phenotypic manifestation varies, ranging from normal, multiple abortions, mental retardation and certain syndromes that are not reported to be solely due to the UPD [Table 2].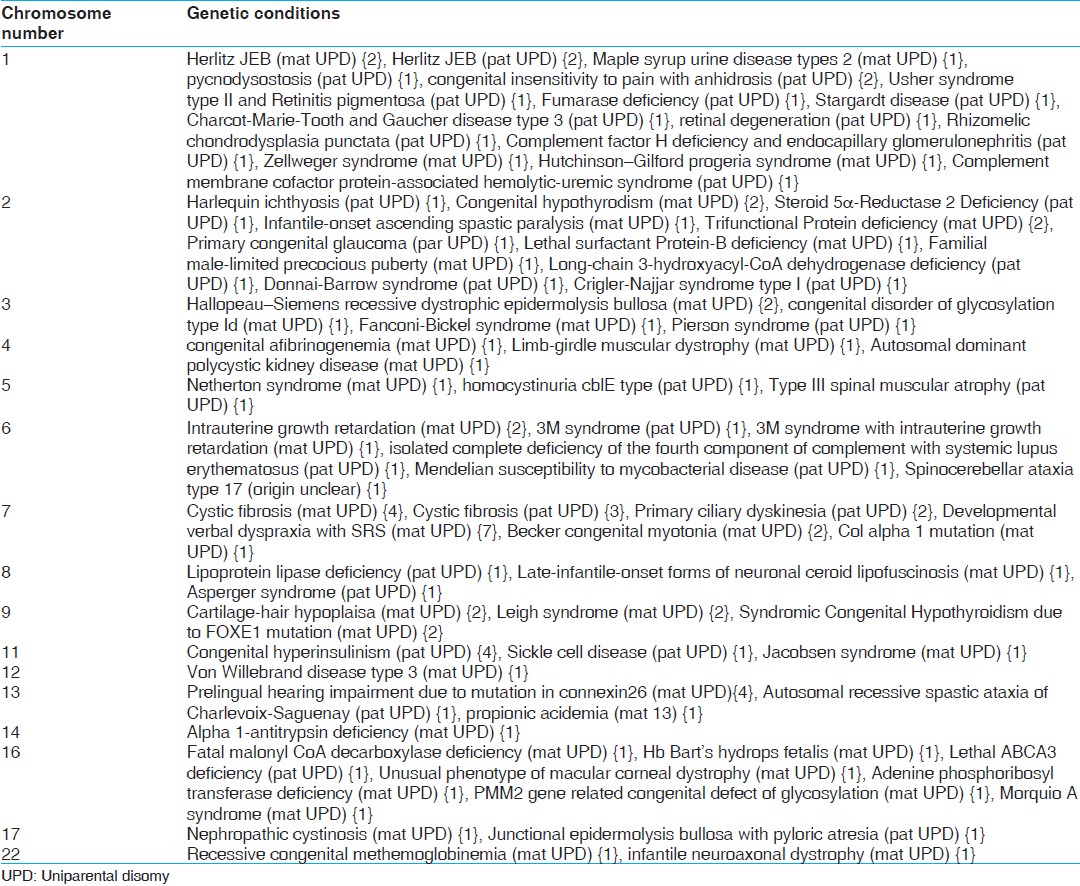 | Table 2: Chromosome wise genetic conditions (number of cases in curly brackets) from the reports of UPD with normal karyotypes
Click here to view |
 Materials and Methods Materials and Methods | |  |
This data mining study was carried out using freely available and regularly updated online database of all published UPD cases. [5] The main focus of this study was to know the extent of genetic conditions that are associated with UPD in addition to those, which are clinically diagnosed in terms of specific chromosomal UPD [Table 1]. The cases with a normal karyotype were selected and those with microscopic chromosomal aberrations were excluded in order to focus on the occult chromosomal aberrations [Figure 1].
 Results Results | |  |
The reports of cases as per the inclusion criteria mentioned above, i.e., more than 170 (out of about 670) were analyzed for the phenotypic expression, chromosome involved in UPD in terms of maternal or paternal origin and related information. In addition to the known UPD related syndromes [Table 1], many other genetic conditions were also associated with UPD [Table 2] in addition to non-specific and unclear phenotypes in about 18 reports, which suggest wide ranging role of UPD. The role of Robertsonian translocations is known in multiple abortions; however, there have also been by chance findings of UPD in cases of bad obstetric history. [6] The presence of UPD in normal individuals detected by chance, for example while paternity testing [7] suggests that the frequency of UPD in human population may not be as rare as perceived. The inheritance pattern of majority of the disease conditions was found to be autosomal recessive as queried using Online Mandelian inheritance in Man.
 Discussion Discussion | |  |
A number of genes are found to be imprinted either for paternal or maternal chromosomal origin. [8] UPD can result in disease mainly if it affects an imprinted gene, i.e., the expression of which is dependent on the parent of origin due to either heterodisomy (inheritance of both the chromosomes from one parent) or isodisomy (inheritance of two copies of the same chromosome from single parent). [9] UPD can also facilitate expression of recessive mutation by loss of wild type allele due to the absence of other parental chromosome. The clinical diagnosis of certain syndromes is based on the detection of UPD [Table 1]; however, there are a number of UPD reports; maternal or paternal [Table 2] in various disease conditions as well as normal individuals detected mainly by chance. A larger population study of whole chromosome or segmental UPD affecting these genes may provide important clues regarding contribution of UPD as a mechanism of pathogenesis in recessive genetic conditions mainly.
Isodisomy of a chromosomal region bearing heterozygous recessive mutation leads to homozygosity and hence disease condition. The data mining exercise indicated that the majority of the disease conditions were due to the isodisomy of autosomal recessive mutations. Prenatal molecular diagnosis in such cases can be suggested if an isodisomy affecting the disease causing recessive mutation is detected. As highlighted by Chediak-Higashi syndrome, in case of this kind of disease causing mechanism imprinting does not play a role. It is just activation of a recessive gene and not related to any imprinting defect!
Summarizing, the range of phenotypes associated with UPD of a chromosome, i.e., maternal or paternal is wide and needs to be comprehensively studied with larger cohorts. Such studies can bring to light the extent of correlation of UPD, genomic imprinting, mechanisms favoring expression of recessive mutations and associated phenotypes. These studies involving whole genome scanning in terms of single nucleotide polymorphism arrays in various unclassified birth defects and even normal population can be informative.
 References References | |  |
| 1. | Engel E. A new genetic concept: Uniparental disomy and its potential effect, isodisomy. Am J Med Genet 1980;6:137-43. 
|
| 2. | Spence JE, Perciaccante RG, Greig GM, Willard HF, Ledbetter DH, Hejtmancik JF, et al. Uniparental disomy as a mechanism for human genetic disease. Am J Hum Genet 1988;42:217-26. 
|
| 3. | Dawson AJ, Chernos J, McGowan-Jordan J, Lavoie J, Shetty S, Steinraths M, et al. CCMG guidelines: Prenatal and postnatal diagnostic testing for uniparental disomy. Clin Genet 2011;79:118-24. 
|
| 4. | Kotzot D, Utermann G. Uniparental disomy (UPD) other than 15: Phenotypes and bibliography updated. Am J Med Genet A 2005;136:287-305. 
|
| 5. | Cases with uniparental disomy. Jena: Institute of Human Genetics. Available from: http://www.fish.uniklinikum-jena.de/UPD.html. [Last accessed on 2012 Jun]. 
|
| 6. | Cases with uniparental disomy. Jena: Institute of Human Genetics. Available from: http://www.fish.uniklinikum-jena.de/UPD/References.html. (No. 37, 58, 101, 108, 183, 237, 348, 450). [Last accessed on 2012 Jun]. 
|
| 7. | Cases with uniparental disomy. Jena: Institute of Human Genetics. Available from: http://www.fish.uniklinikum-jena.de/UPD/References.html. (No. 72, 156, 206, 255, 341, 377, 388, 480, 578). [Last accessed on 2012 Jun]. 
|
| 8. | Jirtle RL. Imprinted genes: By species. Available from: http://www.geneimprint.com. 
|
| 9. | Liehr T. Cytogenetic contribution to uniparental disomy (UPD). Mol Cytogenet 2010;3:8. 
|
[Figure 1]
[Table 1], [Table 2]
|






