|
 
 |
|
ORIGINAL ARTICLE |
|
|
|
| Year : 2013 | Volume
: 19
| Issue : 3 | Page : 301-310 |
| |
Inter and intra ethnic variation of vitamin K epoxide reductase complex and cytochrome P450 4F2 genetic polymorphisms and their prevalence in South Indian population
Dhakchinamoorthi Krishna Kumar1, Deepak Gopal Shewade1, Sajjanavar Manjunath2, Prayaga Ushakiran3, Gangadharan Reneega4, Chandrasekaran Adithan1
1 Department of Pharmacology, Indian Council of Medical Research Centre for Advance Research in Pharmacogenomics, Jawaharlal Institute of Postgraduate Medical Education and Research, Pondicherry, India
2 Department of Pharmacology, M.R Medical College, Gulbarga, Karnataka, India
3 Department of Pharmacology, Rangaraya Medical College, Kakinada, Andhra Pradesh, India
4 Department of Pharmacology, Thiruvananthapuram Medical College, Thiruvananthapuram, Kerala, India
| Date of Web Publication | 30-Oct-2013 |
Correspondence Address:
Dhakchinamoorthi Krishna Kumar
Department of Pharmacology, Indian Council of Medical Research Centre for Advance Research in Pharmacogenomics, Jawaharlal Institute of Postgraduate Medical Education and Research, Pondicherry - 605 006
India
 Source of Support: This research project was funded by Indian Council of Medical Research (ICMR), New Delhi, India, Conflict of Interest: None  | 1 |
DOI: 10.4103/0971-6866.120817

 Abstract Abstract | | |
Background: Genetic variation in the vitamin K epoxide reductase complex (VKORC1) and cytochrome P450 4F2 (CYP4F2) genes were found to be strongly associated with the oral anticoagulant (OA) dose requirement. The distribution of genetic variation in these two genes was found to show large inter- and intra-ethnic difference.
Materials and Methods: A total of 470 unrelated, healthy volunteers of South Indians of either sex (age: 18-60 years) were enrolled for the study. A 5 ml of venous blood was collected and the genomic deoxyribonucleic acid (DNA) was extracted by using phenol-chloroform extraction method. Real-time quantitative polymerase chain reaction (RT-PCR) method was used for genotyping.
Results: The variant allele frequencies of VKORC1 rs2359612 (T), rs8050894 (C), rs9934438 (T) and rs9923231 (A) were found to be 11.0%, 11.8%, 11.7% and 12.0%, respectively. The variant allele VKORC1 rs7294 was (80.1%) more frequent and the variant allele CYP4F2 * 3 was found to be 41.8% in South Indians. The allele, genotype and haplotype frequencies of VKORC1 and CYP4F2 gene were distinct from other compared HapMap populations (P < 0.0001).
Conclusion: The findings of our study provide the basic genetic information for further pharmacogenetic based investigation of OA therapy in the population.
Keywords: Cytochrome P450 4F2, haplotypes, oral anticoagulants, South Indians, vitamin K epoxide reductase complex
How to cite this article:
Kumar DK, Shewade DG, Manjunath S, Ushakiran P, Reneega G, Adithan C. Inter and intra ethnic variation of vitamin K epoxide reductase complex and cytochrome P450 4F2 genetic polymorphisms and their prevalence in South Indian population. Indian J Hum Genet 2013;19:301-10 |
How to cite this URL:
Kumar DK, Shewade DG, Manjunath S, Ushakiran P, Reneega G, Adithan C. Inter and intra ethnic variation of vitamin K epoxide reductase complex and cytochrome P450 4F2 genetic polymorphisms and their prevalence in South Indian population. Indian J Hum Genet [serial online] 2013 [cited 2016 May 24];19:301-10. Available from: http://www.ijhg.com/text.asp?2013/19/3/301/120817 |
 Introduction Introduction | |  |
Vitamin K epoxide reductase (VKOR) enzyme converts vitamin K (VK) 2, 3-epoxide to VK hydroquinone, a required cofactor for the post-translational gamma-carboxylation of several blood coagulation factors (factors II, VII, IX, X, Protein C and S) and other VK-dependent proteins, such as osteocalcin bone Gla protein and matrix Gla protein. [1],[2],[3] The gamma- glutamyl carboxylase enzyme requires reduced VK as a cofactor in the clotting pathway. Oral anticoagulants (OAs) (warfarin, acenocoumarol and phenprocoumon) inhibit the regeneration of reduced VK by targeting the enzyme VKOR and there by blocks the clotting mechanism. [1],[4] The inter-individual variability in OA dose is influenced by the genes of two enzymes, cytochrome P450 (CYP) 2C9, the enzyme that metabolizes the OAs and vitamin K epoxide reductase complex (VKORC1), the pharmacological target enzyme of those drugs. The genetic variation in the CYP2C9 gene (in particular, the common CYP2C9*2 and CYP2C9*3 alleles) was a result in decreased enzyme activity and affects the pharmacokinetics of OA and its dose requirement. The variants in the VKORC1 gene influence the pharmacodynamics response to OAs. [5],[6],[7] Polymorphisms in the VKORC1 gene encoding the VKOR enzyme leads to inter individual variability in susceptibility to develop bleeding and other adverse reactions to oral anti-coagulants. [8],[9] The impact of VKORC1 genetic variation on warfarin dose requirement confirmed that the genetic polymorphisms of VKORC1 are significant determinants of inter- individual dose requirement. It has been found that genetic variations in the CYP2C9 and VKORC1 genes together account for 35-50% variability in the dose requirement for initiation and maintenance of OAs. [5],[10],[11] There is an evidence that specific haplotypes can help to determine the dose of OAs. Single nucleotide polymorphisms (SNPs) found in haplotype A (3730G > A, 2255C > T, 1542G > C, 1173 C > T and − 1639 G > A) versus wild type alleles in haplotype B can be used to determine whether the person require low, intermediate or high dose of OAs. [12]
CYP4F2 genetic polymorphism (rs2108622) was found to be associated with reduced hepatic CYP4F2 enzyme activity and higher levels of VK. The presence of this variant allele is associated with a higher warfarin dose requirement. [13],[14],[15] CYP4F2 functions as a VK mono-oxidase, probably generating the ω-hydroxy derivative of the substrate. Carriers of the CYP4F2 433M allele have a reduced capacity to metabolize VK, secondary to a CYP4F2 dependent decrease in the steady state hepatic concentrations of the enzyme. Therefore, patients with the CYP4F2*3 polymorphism are likely to have an elevated hepatic level of VK and thus are likely to require a higher warfarin dose to prompt the expected anticoagulant response. [16]
South Indians constitute about 21.7% of the total population of India and residing at four states (Tamil Nadu, Andhra Pradesh, Kerala and Karnataka) ( http://www.census india.govt.in/2011census, accessed on August, 2011). This population shares a common ancestry (Dravidians), but the present day population distinguishes themselves from one another in terms of language, culture and dietary habits with limited admixture. Reich et al. [17] found that the ancestral South Indians were distinct from the ancestral North Indians and East Indians.
In our previous studies, we have found that the polymorphic profile of the genes encoding some of the Phase I and Phase II drug metabolizing enzymes (CYP2E1, CYP2A6, CYP3A5, CYP2C9, CYP2C19, CYP2D6, thiopurine methyl transferase and uridine diphosphate-glucoronyl transferases) as well as drug transporters (multidrug resistance 1 and organic cation transporters 1) were significantly different between South Indians and other population groups. [18],[19],[20],[21],[22],[23] These findings raise the possibility that the frequency of variants of other genes including the OA dose determining VKORC1and CYP4F2 genes may be different in our population. This is the first study to establish the haplotype structure of the VKORC1 and genotype/allele frequencies of the CYP4F2 in India, in particular for South Indians. In this study, we have also compared our data with those of other population groups included in the HapMap project ( http://hapmap.ncbi.nlm.nih.gov/ ).
 Materials and Methods Materials and Methods | |  |
Study subjects
The present study included 470 unrelated healthy volunteers from South Indian states (Tamil Nadu, Andhra Pradesh, Kerala and Karnataka) of both genders, aged between 18 and 60 years with a family history of three generations in South India and speaking any one of the South Indian languages. Blood samples were collected from Tamilians (n = 110), Andhrites (n = 120), Kannadigas (n = 120) and Keralites (n = 120). The Institute ethics committee approved the study and was conducted according to declaration of Helsinki. Written informed consent was obtained from all volunteers.
Genotyping procedure
A volume of 5 ml of venous blood was collected using sodium ethylene diamine tetra acetic acid as anticoagulant. Deoxyribonucleic acid (DNA) was extracted by using a standard phenol: chloroform protocol. Genotyping of VKORC1 and CYP4F2 was carried out by real-time thermo cycler (7300 Applied Biosystems; Life Technologies Corporation, Carlsbad, CA, USA) using TaqMan SNP genotyping assays (Assay ID for VKORC1 C__7473918_10, rs7294, C__26291751_10, rs2359612, C__2847860_10, rs8050894, C__30204875_10, rs9934438, C__30996661_30, rs9923231 and assay ID for CYP4F2 C_16179493_40, rs2108622). The polymerase chain reaction (PCR) was carried out in duplicate in a 20-μL final volume that contained 10 μL of TaqMan universal PCR master mix (2x), 0.5 μL of 20x working stock of SNP genotyping assay and 4.5 μL of genomic DNA diluted in DNAase free water and 5 μL of Milli-Q water (Millipore Corporate Headquarters, Billerica, MA, USA). The thermocycler conditions included one cycle at 50°C for 2 min; one cycle at 95°C for 10 min to activate the AmpliTaq Gold polymerase followed by 40 cycles of denaturation at 92°C for 15 s and annealing/extension at 60°C for 1 min. The allelic discrimination was performed using 7300 SDS software version 1.4 (Applied Biosystems; Life Technologies Corporation, Carlsbad, CA, USA).
Statistical analysis
Statistical analysis was performed using the GraphPad InStat 3 software (GraphPad Software Inc., San Diego, CA, USA). Hardy - Weinberg equilibrium was tested by Chi-square test to compare the observed genotype frequencies of South Indian population with the expected genotype frequencies calculated from the observed allele frequencies. Chi-square test was used for comparisons of two different ethnic groups. P <0.05 was considered as statistically significant. Pairwise linkage disequilibrium (LD) pattern and haplotype frequencies were estimated using HAPLOVIEW 4.1 [24] (Daly Lab, Broad Institute, Cambridge, MA02141, USA). All SNPs with minor allele frequencies of 0.01% were excluded and minimum haplotype frequency was set as 1%. Haplotype blocks were defined by using four gametes rule incorporated by analysis in HAPLOVIEW software. Haplotypes were estimated by accelerated expectation-maximization algorithm in HAPLOVIEW. The confidence interval range for LD was set between 0.7 and 0.98. D' values from 0.7 to 1.0 indicates strong LD between the pair of SNPs. Whereas, D' value <0.7 indicates moderate LD and D' value of <0.2 indicates no LD.
 Results Results | |  |
Among the 470 (245 men, 225 women; age [±SD] 27.0 ± 7.9 years) healthy volunteers, 445 samples genotyping was successful and included in further analysis. The frequency distribution of VKORC1 and CYP4F2 genotypes and allele in four South Indian populations was found to be in Hardy - Weinberg equilibrium (4 × 3 contingency table; rs7294-χ² =11.52, P = 0.0734, rs2359612-χ² =12.34, P = 0.0547, rs8050894-χ² =9.85, P = 0.1308, rs9934438-χ² =7.48, P = 0.2781, rs9923231-χ² =8.12, P = 0.2295, rs2108622, χ² =5.845, P = 0.4408).
The overall allele and genotype frequency [Table 1]a, b and [Table 2] was also calculated. The genotype and allele frequency of VKORC1 and CYP4F2 in South Indians was compared with HapMap populations (African ancestry in Southwest USA, Utah residents with Northern and Western European, Han Chinese in Beijing [CHB], Chinese in Metropolitan Denver, Gujarati Indians in Houston [GIH], Texas, Japanese in Tokyo [JPT], Japan, Luhya in Webuye, Kenya, Mexican ancestry in Los Angeles, California, Maasai in Kinyawa, Kenya, Toscans in Italy, Yoruba in Ibadan [YRI], Nigeria).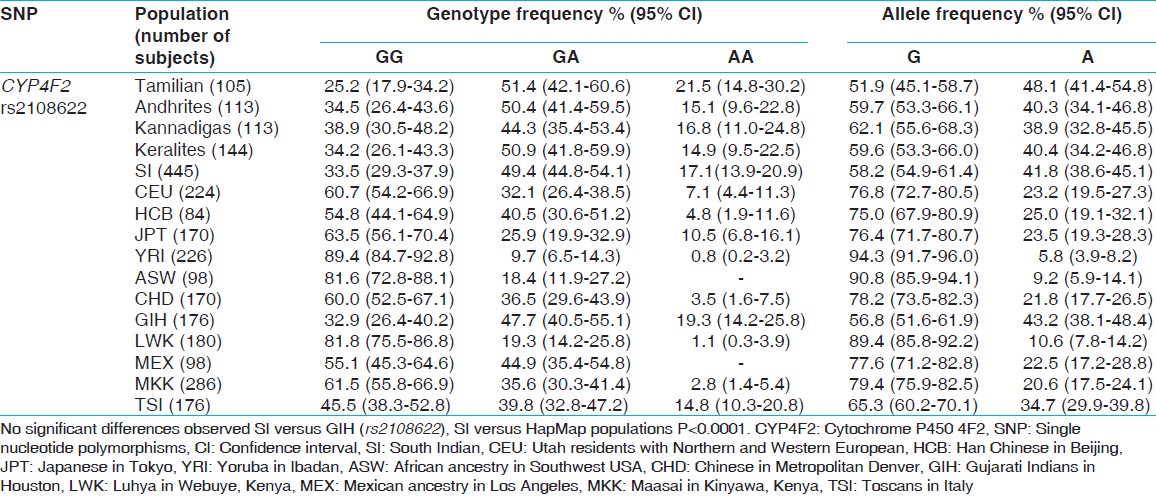 | Table 2: Comparison of genotype and allele frequencies of CYP4F2 gene in SI and other HapMap population
Click here to view |
The haplotype frequencies of VKORC1 in South Indians were compared with major HapMap populations (namely African American, Caucasians, CHB and GIH). Data was obtained from PharmGKB database [Table 3]. The Haplotype structure and LD pattern of VKORC1 in South Indian was compared with the HapMap population [Figure 1].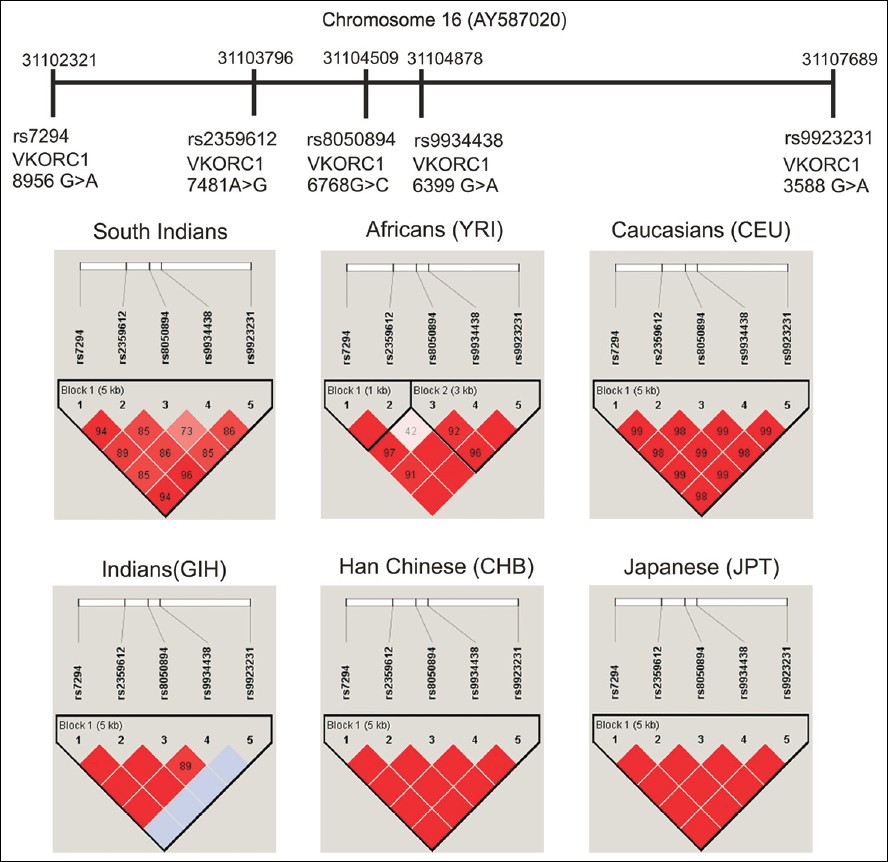 | Figure 1: Linkage disequilibrium pattern of vitamin K epoxide reductase complex genetic variants in south Indians and other HapMap populations (The single nucleotide polymorphisms in Chromosome 16 were positioned according to the order and orientation. Each of the variants is given with its specific chromosomal position and the rsID. Red and pink colors represent a very strong LD pattern (D' >0.8) and white color represents moderate to low LD (D' <0.8 to >0.5). The blue color in Indian (GIH) indicates missing genotype data)
Click here to view |
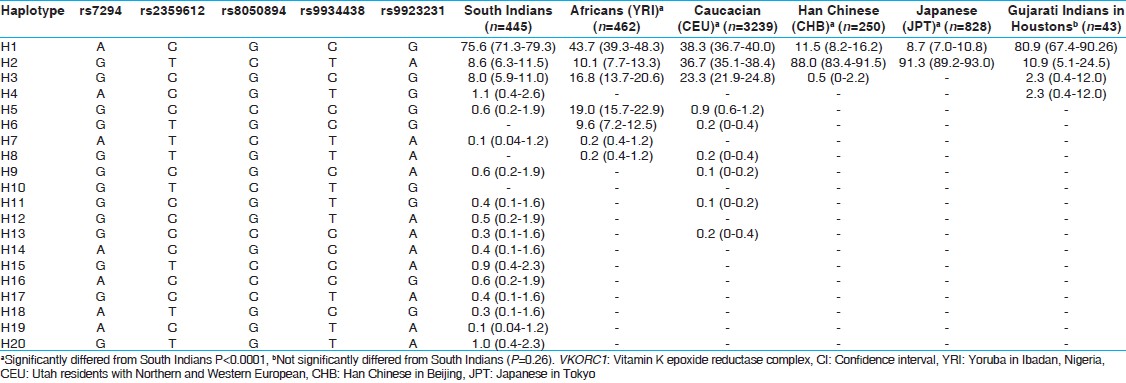 | Table 3: Haplotype frequencies (% [95% CI]) of VKORC1 gene in South Indians and other major HapMap population
Click here to view |
In South Indian population, a strong LD pattern (D' >0.8) was observed between all the SNPs except the LD pattern between rs8050894 and rs9934438 only moderate LD (D' <0.8) was observed. In Africans (YRI), a very strong LD pattern (D' =1) was observed between rs7294 and rs2359612, rs2359612 and rs9934438, rs2359612 and rs9923231, rs7294 and rs9923231 rs9934438 and rs9923231, rs7294 and rs9934438. Only low LD (D' <0.5) was observed between rs2359612 and rs8050894. In Caucasians, all the SNPs were in strong LD (D' >0.8). In CHB and JPT strong LD pattern (D' =1) was observed between all the SNPs. In GIH for rs9923231 genotype data was not available for many samples, but strong LD pattern (D' >0.8) was observed between other SNPs. In the four South Indian populations, the LD pattern was observed. In Tamilians only moderate LD pattern (D' <0.8 to > 0.5) was observed between rs7294 and rs9934438, rs8050894 and rs9934438, rs9934438 and rs9923231. In Andhrites, a moderate LD pattern (D' <0.8) was observed between rs2359612 and rs8050894, rs8050894 and rs9934438, rs8050894 and rs9923231. Strong LD pattern was observed between other SNPs. In Kannadigas and Keralites, a strong LD pattern was observed between all the SNPs. Allele frequencies of most studied SNPs (CYP2C9*2 [rs1799852], CYP2C9*3 [rs1057910], VKORC1 [rs9923231, rs7294, rs9934438] and CYP4F2 [rs2108622]) in South Indians and other HapMap populations were compared [Figure 2].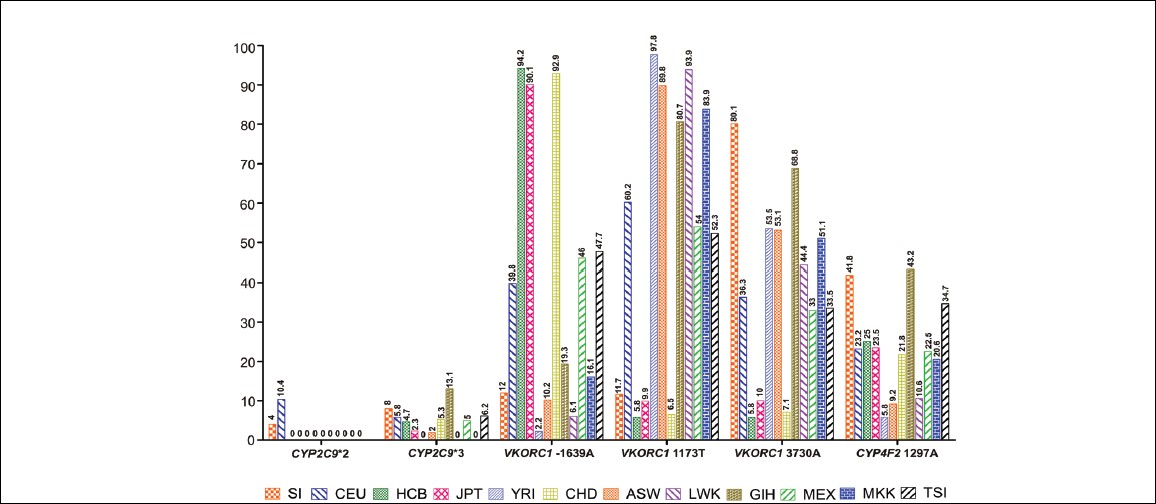 | Figure 2: Allele frequencies of most studied single nucleotide polymorphisms (cytochrome P450 2C9 [CYP2C9]*2 [rs1799852], CYP2C9 * 3 [rs1057910], Vitamin K epoxide reductase complex [rs9923231, rs7294, rs9934438] and CYP4F2 [rs2108622]) genes in South Indians and other HapMap populations
Click here to view |
 Discussion Discussion | |  |
The frequencies of genetic variations may vary between populations as highlighted by several reports from different populations and from the HapMap projects. [25],[26] Hence, it is useful to have the population distribution profile of the genetic polymorphisms and LD pattern in the study population prior to genetic association studies. The present study is the first to report the genotype, allele and haplotype distribution and LD pattern of VKORC1 and CYP4F2 genes in South Indian population with a large sample size. A significant difference in genotype, allele and Haplotype distribution was observed between the present study groups and other populations.
It is well-documented that the two alleles CYP2C9*2 (rs1799852) and CYP2C9*3 (rs1057910) were significantly associated with decreased warfarin dose requirements (warfarin sensitivity) and higher susceptibility to overdose. [27] A direct association of CYP2C9 genotype and anticoagulation status and bleeding was first reported by Higashi et al. [28] Furthermore, the allele frequencies of CYP2C9*2 and CYP2C9*3 diverge considerably among different ethnic groups. [29],[30] In our previous study, we have established the genotype and allele frequency of CYP2C9*2 and CYP2C9*3 in South Indian population. [19]
In the present study, we have focused on the VKORC1 and CYP4F2 genes. Apart from SNPs in CYP2C9 gene and SNPs in the VKORC1 has been also correlated with warfarin dose requirement. [12],[31] The rs9934438 and rs9923231 polymorphisms are most commonly studied and associated with a lower warfarin dose requirement. [32],[33],[34] Another common SNP, the 3730 G > A (rs7294), located in the 3' untranslated region of the gene, is associated with increased warfarin dose requirements (warfarin resistance). [12],[35] In our study, we found that the rs7294 variant was highly frequent (80.1%).
Rieder et al. [12] conducted the haplotype analysis and established a significant contribution of VKORC1 to inter individual variability in warfarin dose. The ten most common SNPs (at positions 381, 861, 2653, 3673, 5808, 6009, 6484, 6853, 7566 and 9041 of the VKORC1 reference sequence [GenBank accession number AY587020]) were used to construct the five major haplotypes and the association of these haplotypes were observed in Caucasian patients. A low dose haplotype group A and high dose haplotype group B were identified. These haplotype frequencies also vary among the different ethnic populations. Haplotype A is more frequent in Asians (89%); whereas in Caucasians, haplotype B is more frequent (58%). [12]
The previous pharmacogenetics studies have provided the indications that the South Indian population may have distinct polymorphic distribution features regarding the VKORC1 locus. In our study, we did not observe a significant difference in the genotyping frequencies of Tamilians, Kannadigas, Andhrites and Keralites. Only the genotype frequencies of rs7294 and rs2359612 in Kannadigas were significantly different from the Tamilians population. The pooled results of South Indians genotypes were compared with that of the HapMap population. We have observed a highly significant difference between the different ethnic population groups and South Indians. Only the Japanese population did not significantly differ for the SNPs rs7294, rs2359612 and rs9934438.
The haplotype frequencies of the VKORC1 locus in South Indians were compared with those of the HapMap population ( http://hapmap.ncbi.nlm.nih.gov/ ) by establishing the haplotype structure for Africans, Caucasians, Han Chinese, Japanese and Indian groups. The haplotype frequencies in South Indians were significantly different from Africans, Han Chineses and Japaneses, but not from the GIH. The major Haplotype observed in our population is ACGCG (75.6%), but this haplotype was observed at different levels in Africans (43.7%), Caucasians (38.3%), Han Chineses (11.5%) and Japaneses (8.7%), but close to that of GIH (80.9%). Thus, the present study reveals that the South Indians are anticipated to fall under the high dose requirement groups for the OAs.
A previous study has established the haplotype frequency (rs7196161, rs17880887, rs9923231, rs2884737, rs9934438 and rs17880624) in the Malaysian Indians and found that the TCGTCA (H7) is more frequent in Indians as compared with Chinese and Malays. The H7 haplotype group is associated with the higher dose requirement. [36] Our study on South Indians is in line with this observation.
In addition to the CYP2C9 and VKORC1 polymorphisms, a SNP in the CYP4F2 gene has been shown to contribute to warfarin dose requirement, albeit to a lesser extent, through genome wide association studies. [14] Another study has shown that patients with a variant allele namely CYP4F2*3, require higher maintenance warfarin dose as compared with those with wild type allele in Han Chinese patients. [33] In this study, we have established the frequency of CYP4F2*3 allele and compared it with that of the HapMap populations. Both the allele and genotype frequencies significantly differed across these population groups with the exception of Gujarati Indians from Houston.
The United States Food and Drug Administration updated the label of warfarin twice: In 2007 advising physicians to consider the use of "genetic tests to improve their initial estimate" of the initial dosage, then in 2010 adding a new table with the range of expected therapeutic warfarin doses based on CYP2C9 and VKORC1 genotypes. Based on the evidences many studies have proposed the algorithms for calculating the maintenance dose and initial dose of OAs using the multivariate statistical techniques. [11],[37],[38],[39],[40],[41],[42],[43],[44] However, the proposed algorithms appear to be specific for each population group very likely due to differences in distribution of polymorphisms.
Thus, it is useful to establish the polymorphic profile of the study population/cohort before conducting pharmacogenetic studies on OAs. At this end, we have established the genotype, allele and haplotype frequencies of VKORC1 and CYP4F2 genotype and allele frequencies for the South Indian population.
 Conclusion Conclusion | |  |
The frequencies of the VKORC1 and CYP4F2 allele/genotype in the South Indian population were distinct from other world population groups. These results will not only contribute to the better understanding of the genetic basis of ethnic variation in OA dose requirement response, but also establishes the frame work for future algorithm based dose determination of OAs in Indians.
 Acknowledgments Acknowledgments | |  |
This research project was funded by Indian Council of Medical Research (ICMR), New Delhi, India. (ICMR Ref. No. 53/17/2003-BMS dated 12/03/2007). Ms. G. Saraswathi, Ms. S. Kalaivani and Mrs. N. Revathy, Technical Assistants, are gratefully acknowledged.
 References References | |  |
| 1. | Presnell SR, Stafford DW. The vitamin K-dependent carboxylase. Thromb Haemost 2002;87:937-46. 
|
| 2. | Tie J, Wu SM, Jin D, Nicchitta CV, Stafford DW. A topological study of the human gamma-glutamyl carboxylase. Blood 2000;96:973-8. 
|
| 3. | Wu SM, Cheung WF, Frazier D, Stafford DW. Cloning and expression of the cDNA for human gamma-glutamyl carboxylase. Science 1991;254:1634-6. 
|
| 4. | Larson AE, Friedman PA, Suttie JW. Vitamin K-dependent carboxylase. Stoichiometry of carboxylation and vitamin K 2,3-epoxide formation. J Biol Chem 1981;256:11032-5. 
|
| 5. | Schalekamp T, Brassé BP, Roijers JF, van Meegen E, van der Meer FJ, van Wijk EM, et al. VKORC1 and CYP2C9 genotypes and phenprocoumon anticoagulation status: Interaction between both genotypes affects dose requirement. Clin Pharmacol Ther 2007;81:185-93. 
|
| 6. | Stehle S, Kirchheiner J, Lazar A, Fuhr U. Pharmacogenetics of oral anticoagulants: A basis for dose individualization. Clin Pharmacokinet 2008;47:565-94. 
|
| 7. | Wadelius M, Chen LY, Eriksson N, Bumpstead S, Ghori J, Wadelius C, et al. Association of warfarin dose with genes involved in its action and metabolism. Hum Genet 2007;121:23-34. 
|
| 8. | Bodin L, Verstuyft C, Tregouet DA, Robert A, Dubert L, Funck-Brentano C, et al. Cytochrome P450 2C9 (CYP2C9) and vitamin K epoxide reductase (VKORC1) genotypes as determinants of acenocoumarol sensitivity. Blood 2005;106:135-40. 
|
| 9. | Rost S, Fregin A, Ivaskevicius V, Conzelmann E, Hörtnagel K, Pelz HJ, et al. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature 2004;427:537-41. 
|
| 10. | Bodin L, Perdu J, Diry M, Horellou MH, Loriot MA. Multiple genetic alterations in vitamin K epoxide reductase complex subunit 1 gene (VKORC1) can explain the high dose requirement during oral anticoagulation in humans. J Thromb Haemost 2008;6:1436-9. 
|
| 11. | Wadelius M, Chen LY, Lindh JD, Eriksson N, Ghori MJ, Bumpstead S, et al. The largest prospective warfarin-treated cohort supports genetic forecasting. Blood 2009;113:784-92. 
|
| 12. | Rieder MJ, Reiner AP, Gage BF, Nickerson DA, Eby CS, McLeod HL, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med 2005;352:2285-93. 
|
| 13. | Singh O, Sandanaraj E, Subramanian K, Lee LH, Chowbay B. Influence of CYP4F2 rs2108622 (V433M) on warfarin dose requirement in Asian patients. Drug Metab Pharmacokinet 2011;26:130-6. 
|
| 14. | Caldwell MD, Awad T, Johnson JA, Gage BF, Falkowski M, Gardina P, et al. CYP4F2 genetic variant alters required warfarin dose. Blood 2008;111:4106-12. 
|
| 15. | Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N, et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet 2009;5:e1000433. 
|
| 16. | McDonald MG, Rieder MJ, Nakano M, Hsia CK, Rettie AE. CYP4F2 is a vitamin K1 oxidase: An explanation for altered warfarin dose in carriers of the V433M variant. Mol Pharmacol 2009;75:1337-46. 
|
| 17. | Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature 2009;461:489-94. 
|
| 18. | Abraham BK, Adithan C, Kiran PU, Asad M, Koumaravelou K. Genetic polymorphism of CYP2D6 in Karnataka and Andhra Pradesh population in India. Acta Pharmacol Sin 2000;21:494-8. 
|
| 19. | Naveen AT, Adithan C, Soya SS, Gerard N, Krishnamoorthy R. CYP2D6 genetic polymorphism in South Indian populations. Biol Pharm Bull 2006;29:1655-8. 
|
| 20. | Jose R, Chandrasekaran A, Sam SS, Gerard N, Chanolean S, Abraham BK, et al. CYP2C9 and CYP2C19 genetic polymorphisms: Frequencies in the south Indian population. Fundam Clin Pharmacol 2005;19:101-5. 
|
| 21. | Arun Kumar AS, Chakradhara Rao US, Umamaheswaran G, Ramu P, Kesavan R, Shewade DG, et al. Haplotype structures of common variants of CYP2C8, CYP2C9, and ADRB1 genes in a South Indian population. Genet Test Mol Biomarkers 2011;15:407-13. 
|
| 22. | Krishnakumar D, Gurusamy U, Dhandapani K, Surendiran A, Baghel R, Kukreti R, et al. Genetic polymorphisms of drug-metabolizing phase I enzymes CYP2E1, CYP2A6 and CYP3A5 in South Indian population. Fundam Clin Pharmacol 2012;26:295-306. 
|
| 23. | Umamaheswaran G, Krishna Kumar D, Kayathiri D, Rajan S, Shewade DG, Dkhar SA, et al. Inter and intra-ethnic differences in the distribution of the molecular variants of TPMT, UGT1A1 and MDR1 genes in the South Indian population. Mol Biol Rep 2012;39:6343-51. 
|
| 24. | Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263-5. 
|
| 25. | Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly P. A haplotype map of human genome. Nature 2005;437:1299-320. 
|
| 26. | Sawyer SL, Mukherjee N, Pakstis AJ, Feuk L, Kidd JR, Brookes AJ, et al. Linkage disequilibrium patterns vary substantially among populations. Eur J Hum Genet 2005;13:677-86. 
|
| 27. | Margaglione M, Colaizzo D, D'Andrea G, Brancaccio V, Ciampa A, Grandone E, et al. Genetic modulation of oral anticoagulation with warfarin. Thromb Haemost 2000;84:775-8. 
|
| 28. | Higashi MK, Veenstra DL, Kondo LM, Wittkowsky AK, Srinouanprachanh SL, Farin FM, et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA 2002;287:1690-8. 
|
| 29. | Stubbins MJ, Harries LW, Smith G, Tarbit MH, Wolf CR. Genetic analysis of the human cytochrome P450 CYP2C9 locus. Pharmacogenetics 1996;6:429-39. 
|
| 30. | Scott SA, Khasawneh R, Peter I, Kornreich R, Desnick RJ. Combined CYP2C9, VKORC1 and CYP4F2 frequencies among racial and ethnic groups. Pharmacogenomics 2010;11:781-91. 
|
| 31. | Veenstra DL, You JH, Rieder MJ, Farin FM, Wilkerson HW, Blough DK, et al. Association of Vitamin K epoxide reductase complex1 (VKORC1) variants with warfarin dose in a Hong Kong Chinese patient population. Pharmacogenet Genomics 2005;15:687-91. 
|
| 32. | Dumas TE, Hawke RL, Lee CR. Warfarin dosing and the promise of pharmacogenomics. Curr Clin Pharmacol 2007;2:11-2. 
|
| 33. | Liang R, Li L, Li C, Gao Y, Liu W, Hu D, et al. Impact of CYP2C9*3, VKORC1-1639, CYP4F2 rs2108622 genetic polymorphism and clinical factors on warfarin maintenance dose in Han-Chinese patients. J Thromb Thrombolysis 2012;34:120-5. 
|
| 34. | El Din MS, Amin DG, Ragab SB, Ashour EE, Mohamed MH, Mohamed AM. Frequency of VKORC1 (C1173T) and CYP2C9 genetic polymorphisms in Egyptians and their influence on warfarin maintenance dose: Proposal for a new dosing regimen. Int J Lab Hematol 2012;34:517-24. 
|
| 35. | Wells PS, Holbrook AM, Crowther NR, Hirsh J. Interactions of warfarin with drugs and food. Ann Intern Med 1994;121:676-83. 
|
| 36. | Lal S, Sandanaraj E, Jada SR, Kong MC, Lee LH, Goh BC, et al. Influence of APOE genotypes and VKORC1 haplotypes on warfarin dose requirements in Asian patients. Br J Clin Pharmacol 2008;65:260-4. 
|
| 37. | Anderson JL, Horne BD, Stevens SM, Grove AS, Barton S, Nicholas ZP, et al. Randomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anticoagulation. Circulation 2007;116:2563-70. 
|
| 38. | Pavani A, Naushad SM, Rupasree Y, Kumar TR, Malempati AR, Pinjala RK, et al. Optimization of warfarin dose by population-specific pharmacogenomic algorithm. Pharmacogenomics J 2012;12:306-11. 
|
| 39. | Gage BF, Lesko LJ. Pharmacogenetics of warfarin: Regulatory, scientific, and clinical issues. J Thromb Thrombolysis 2008;25:45-51. 
|
| 40. | International Warfarin Pharmacogenetics Consortium, Klein TE, Altman RB, Eriksson N, Gage BF, Kimmel SE, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med 2009;360:753-64. 
|
| 41. | Perini JA, Struchiner CJ, Silva-Assunção E, Santana IS, Rangel F, Ojopi EB, et al. Pharmacogenetics of warfarin: Development of a dosing algorithm for brazilian patients. Clin Pharmacol Ther 2008;84:722-8. 
|
| 42. | Sconce EA, Khan TI, Wynne HA, Avery P, Monkhouse L, King BP, et al. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: Proposal for a new dosing regimen. Blood 2005;106:2329-33. 
|
| 43. | Schelleman H, Chen J, Chen Z, Christie J, Newcomb CW, Brensinger CM, et al. Dosing algorithms to predict warfarin maintenance dose in Caucasians and African Americans. Clin Pharmacol Ther 2008;84:332-9. 
|
| 44. | Zhu Y, Shennan M, Reynolds KK, Johnson NA, Herrnberger MR, Valdes R Jr, et al. Estimation of warfarin maintenance dose based on VKORC1 (−1639 G>A) and CYP2C9 genotypes. Clin Chem 2007;53:1199-205. 
|
[Figure 1], [Figure 2]
[Table 1], [Table 2], [Table 3]
| This article has been cited by | | 1 |
Warfarin pharmacogenetics: How close are we to clinical practice? |
|
| Gaikwad, T., Shetty, S., Ghosh, K. | | Indian Journal of Human Genetics. 2013; 19(3): 277-278 | | [Pubmed] | |
|
 |
|






