|
 
 |
|
ORIGINAL ARTICLE |
|
|
|
| Year : 2013 | Volume
: 19
| Issue : 4 | Page : 430-436 |
| |
Promoter variants in interleukin-6 and tumor necrosis factor alpha and risk of coronary artery disease in a population from Western India
Aparna A Bhanushali, BR Das
Research and Development, Super Religare Laboratories Ltd., Goregaon West, Mumbai, Maharashtra, India
| Date of Web Publication | 4-Jan-2014 |
Correspondence Address:
B R Das
Research and Development, SRL Ltd., Prime Square Building, Plot No. 1, S. V. Road, Goregaon West, Mumbai - 400 062, Maharashtra
India
 Source of Support: None, Conflict of Interest: None
DOI: 10.4103/0971-6866.124371

 Abstract Abstract | | |
Introduction: A central component of the atherosclerotic process is inflammation. Single nucleotide polymorphisms (SNPs) present in the promoter region of various cytokines can lead to altered levels of the transcript and a state of low-grade inflammation exacerbating the risk of coronary artery disease (CAD). The present work tries to understand the role of permissive promoter variants in the interleukin-6 gene (IL-6-174G/C) and the tumor necrosis factor alpha (TNFα-308G/A) in the causation of CAD and also dyslipidemia.
Materials and Methods: Genotyping was conducted on 100 cases of CAD and 150 controls by the allele termination assay SNaPshot. Biochemical parameters were determined by routine enzymatic endpoint methods. The results were analyzed by appropriate statistical methods.
Results: No differences in the minor allele frequency IL-6-174G/C SNP were seen between cases and controls (0.13 vs. 0.12). The differences in the allele frequency of TNFα-308A between cases (6%) and controls (2%) have led to an odds ratio, 3.370; 95% confidence interval, 1.039-11.543; P=0.033 in the univariate analysis. In the final logistic regression analysis, however none of the variants were associated with an increased risk of CAD.
Conclusions: In summary, no association of the permissive promoter variants in the IL-6 gene and the TNFα gene were seen with an increased CAD risk. These and other studies highlight the importance of doing population specific studies.
Keywords: Association study, coronary artery disease, India, interleukin-6, SNaPshot, single nucleotide polymorphisms, tumor necrosis factor alpha
How to cite this article:
Bhanushali AA, Das B R. Promoter variants in interleukin-6 and tumor necrosis factor alpha and risk of coronary artery disease in a population from Western India. Indian J Hum Genet 2013;19:430-6 |
How to cite this URL:
Bhanushali AA, Das B R. Promoter variants in interleukin-6 and tumor necrosis factor alpha and risk of coronary artery disease in a population from Western India. Indian J Hum Genet [serial online] 2013 [cited 2016 May 24];19:430-6. Available from: http://www.ijhg.com/text.asp?2013/19/4/430/124371 |
 Introduction Introduction | |  |
Over the last decade, there has been a consensus that inflammatory processes underlie atherosclerosis. [1],[2] Inflammation acts at the initial stages of atherosclerosis, where the presence of modified low density lipoprotein (LDL) particles induces a number of specialized cytokines, which result in directed migration of the monocytes to the intimal layer. [3],[4] In the later stages inflammation precipitates adverse clinical events. [5]
Since the levels of circulating cytokines vary considerably inter-individually, genetic variants have been implicated. Until date, many single nucleotide polymorphisms (SNPs) influencing the variance of lipid and proinflammatory molecules have been identified both by candidate-gene as well as genome wide association studies. [6],[7],[8],[9] SNPs in the promoter region of cytokine genes have garnered attention since the presence of the distinct allele is often associated with the baseline cytokine levels and/or also with the response to insult. [10],[11],[12] Thus, these permissive promoter SNPs can lead to the state of low-grade chronic inflammation exacerbating the risk of coronary artery disease (CAD). [13]
The interleukin-6 (IL-6)-174G/C is one such well characterized polymorphism in the promoter [14] that affects the rate of IL-6 serum levels and IL-6 gene transcription. [15] IL-6 is the only cytokine that can stimulate the synthesis of all those involved in the inflammatory response: C-reactive protein, serum amyloid A, fibrinogen, α1-antichymotrypsin and haptoglobin. [16]
Another promoter polymorphism in the tumor necrosis factor alpha (TNFα) gene-308G/A polymorphism has been associated with an increased production of TNFα, with the uncommon A allele shown to be a significantly stronger activator of transcription in vitro. [17],[18] TNFα, is a cytokine with a wide range of proinflammatory activities and it occupies a pivotal role in the initiation and amplification of the inflammatory cascade in clinical conditions such as rheumatoid arthritis and atherogenesis. [19],[20]
With India gearing to develop the highest incidence of CAD, it is important to evaluate the new markers especially genetic variants in our population. It is necessary to determine their frequency, since it can translate into different risk related incidence for the population. In the present study, permissive promoter variants in (IL-6-174G > C) and (TNFα-308G > A) have been evaluated and their association with CAD risk and lipid levels determined in the select Indian population (Indo-Europeans).
 Materials and Methods Materials and Methods | |  |
Subjects
The study was performed on a total of 250 unrelated individuals which consisted of 100 patients with CAD confirmed by coronary angiography: >50% stenosis in one or more arteries and stable or unstable angina and 150 controls: Examined clinically and investigated by electrocardiography to exclude CAD. This cohort consisted of subjects of Indo European descent from Maharashtra in Western India. Informed consent was obtained from all the subjects. The study is in accordance with the Helsinki declaration and was approved by the local ethical committee. A detailed case record form pertaining to information on demographics, medical history and coronary risk factors such as the presence of diabetes, hypertension, smoking, life-style and current medication was completed for each participant through personal interviews and through perusal of their medical records.
Blood specimens were collected after an overnight fast of 12 h by venipuncture using the vacutainer system from Becton Dickinson (Franklin Lakes, NJ USA) in the anti-coagulant ethylenediaminetetraacetic acid (EDTA) as well as plain bulb for serum. Serum, EDTA plasma samples were separated by centrifugation and aliquots were preserved at-20°C until analysis.
Biochemical parameters
The laboratory parameters namely serum total cholesterol (TC) triglyceride (TG) and high density lipoprotein-cholesterol (HDL-C) levels were determined by routine enzymatic endpoint methods (X Imola; Randox Laboratories Ltd. UK). LDL-C and very low-density lipoprotein cholesterol (VLDL-C) were calculated according to Friedewald's formula.
Deoxyribonucleic acid extraction
Genomic DNA was isolated from peripheral blood using QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). DNA yield and purity was determined by measuring absorbance at 260/280 nm.
SNP genotyping
Polymorphisms IL-6-174G > C (rsid1800795), TNF-308G > A (rsid1800629) were determined by a variation of the allele termination assay (ABI PRISM® SNaPshot™ ). The chemistry of this assay is based on the dideoxy single-base extension of an unlabeled oligonucleotide primer. The polymerase extends the specific SNaPshot primer by one nucleotide, adding a single ddNTP to its 3'-end.
Using the contig sequence specific to the SNP, SNaPshot primer was designed to end one base before the SNP. The length of the primer was modified by the addition of non-homologous tails, poly (dGACT) added at the 5′-end. The SNaPshot assay was performed in the following manner. Briefly, 25 μl PCR reaction was carried out using the specific PCR primers and annealing temperature of 58°C for 30 cycles. The PCR products were then purified by a combined Exonuclease (ExoI) and shrimp alkaline phosphatase (SAP) treatment where 5 μl of ExoSAP-IT was added to 15 μl of post-PCR reaction product. The mixture was incubated at 37°C for 15 min and then inactivated by heating at 80°C for 15 min. The specific SNaPshot primer was reconstituted to give a final working dilution of 0.2 μM, the SNaPshot reaction contained 5 μl of 2 × multiplex ready reaction mix, 1 μl of the SNaPshot primer, 1 μl PCR product and 3 μl of de-ionized water in a 10 μl reaction. The reaction mixture was centrifuged briefly and subjected to 25 cycles of 96°C for 10 s, 50°C for 5 s and 60°C for 30 s. Post SNaPshot PCR, the products were again purified, by the alcohol purification method. The purified product was reconstituted in 10 μl of de-ionized water. Subsequently, 0.5 μl of the SNaPshot product, 0.5 μl GeneScan-120 LIZ size standard along with 9 μl Hi-Di formamide was added to each tube, denatured at 95°C for 5 min, kept on ice, transferred into sequencing plate. Capillary electrophoresis was then undertaken on the automated ABI prism 3100 Avant genetic analyzer. The data was analyzed using GeneMapper™ 4.0 Software. The fragment length and color of the alleles obtained is indicated below. 10% of random samples were also sequenced using PCR primers to check concordance with the SNaPshot results.
Statistical analysis
Allele frequency was calculated as the number of occurrences of the test allele in the population divided by the total number of alleles. Any deviation of the genotype frequencies from the Hardy-Weinberg equilibrium (HWE) was assessed by Fischer's exact test. Chi-square tests were used for comparison of binary variables across groups. To determine the risk for CAD, odds ratio (OR) was determined by both univariate analysis, as well as multivariate analysis adjusting for other co-variables. Routine statistical analysis were carried out with the SPSS version 15 software (SPSS Inc., Chicago, IL).
 Results Results | |  |
[Table 1] displays the means and standard deviations (SDs) for the study subjects for relevant biochemical characteristics as well as risk factors. The mean age of CAD incidence was 48 years (SD ± 11). The mean age of controls was 50 years (SD ± 11). It was seen that the cases showed a higher incidence of the traditional risk factors associated with CAD such as smoking, diabetes, hypertension and presence of family history. The genotype and allele frequencies have been enumerated in [Table 2]. The genotypes were identified by fragment analysis as indicated in [Figure 1] along with the corroborating sequencing electropherogram. There was no significant departure from the HWE determined by the exact test (P < 0.05). No discordance could be detected for samples analyzed twice and the success rate of genotyping was ~ 99%. In the present study, no significant differences were observed in the frequency of the IL-6-174G > C genotypes between cases: GG, 77%; GC, 20%; CC 3% and controls GG, 80%; GC, 17%; CC, 3%. On the other hand, the genotype frequency of TNF-308G > A in cases was: GG, 89%, GA, 10%; AA, 1% and in controls was: GG, 97%; GA, 3%; AA, 0%. The differences in the minor allele frequency-308A between cases and controls have led to an OR, 3.370; 95% confidence interval (CI), 1.039-11.543; P, 0.033 in the univariate analysis. The effect of the variants on plasma lipid levels was assessed by comparing the mean concentration of the lipids based on genotypes and performing appropriate statistical analysis. The genotype-phenotype correlation was done only on controls as most of the cases were on lipid lowering medication. No association of the genotypes with any of the lipid levels were seen as indicated in [Table 3]. To determine if the variants are associated with an increased risk of CAD stepwise logistic regression analysis, which included nine variables was performed [Table 4]. In the final model hypertension (OR: 5.732, 95% CI: 2.194-14.970, P < 0.0001) and smoking (OR: 5.027, 95% CI: 1.908-13.243, P, 0.001) were associated with an increased risk of CAD, whereas alcohol consumption seemed to have a protective effect (OR: 0.272, 95% CI: 0.075-0.990, P, 0.048). The TNF-308G > A as a risk variable was removed on step 5 of the model (OR: 2.023, 95% CI: 0.358-11.426-14.970, P, 0.425) and IL-6-176G > C genotype on step 8 of the final model (OR: 1.598, 95% CI: 0.693-3.685, P, 0.271).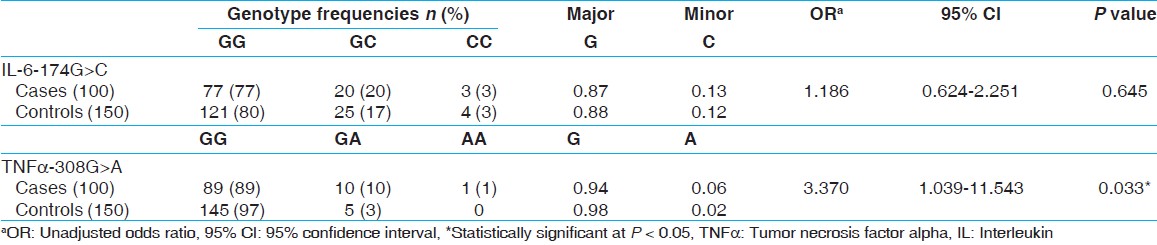 | Table 2: Genotype and allele frequencies of inflammation related variants
Click here to view |
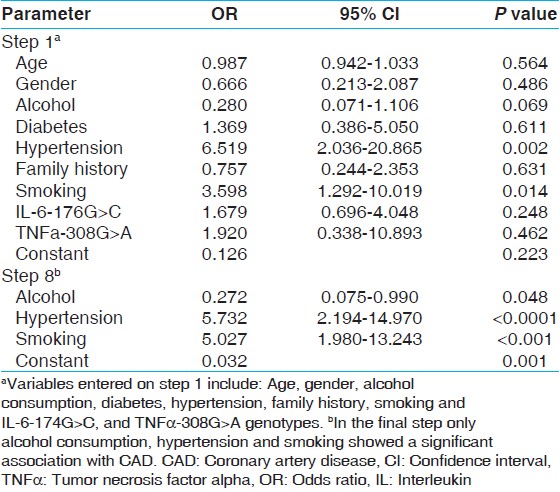 | Table 4: Stepwise backward logistic regression for inflammation related variants in IL-6, IL-10 and TNFα genes association with CAD risk
Click here to view |
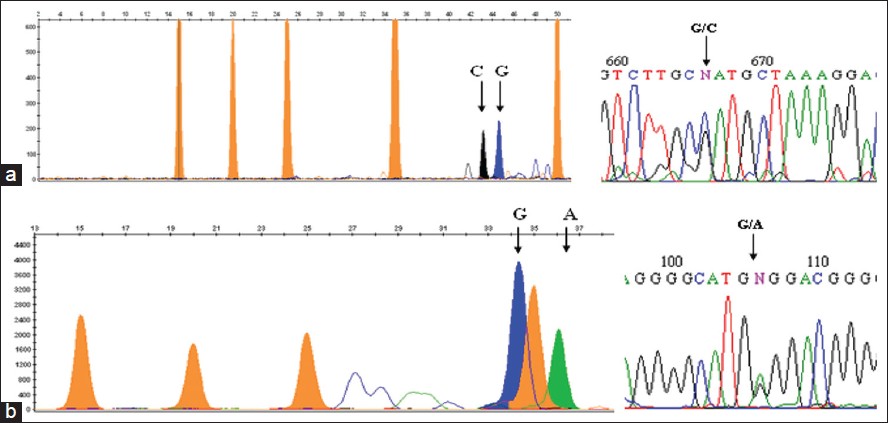 | Figure 1: Genotypes have been shown by SNaPshot assay. The genotypes are analysed based on the color of the peak and fragment length. The orange peaks indicate size standard LIZ120GS. (a) Interleukin - 6 - 174G>C genotyping by SNaPshot indicates both G and C allele. The corroborating sequencing electropherogram is adjacent. (b) Tumor necrosis factor alpha - 308G>A genotyping indicates both G and A allele. The sequencing electropherogram of the same is adjacent
Click here to view |
 Discussion Discussion | |  |
Inflammation is one of the hallmarks of atherosclerosis and hence several studies have evaluated the utility of inflammatory biomarkers in predicting cardiovascular disease. [21] In the present study, two permissive promoter variants-174G/C in the IL-6 gene and-308G/A in the TNFα have been evaluated with respect to alterations in lipid metabolism and CAD risk in our population.
There is a considerable interethnic variation in the allelic distribution of IL-6 G > C polymorphism, with a C allele frequency of 0.41 in Caucasians, [14] 0.15 in Gujarati Indians in US and 0.05 in Afro-Caribbean's. [15] The minor allele frequency-174 (C) was relatively high in the Finnish population and varied between 0.53 and 0.56 in different studies. [22],[23],[24] In the current study, the minor allele frequency-174 (C) was 0.12 in controls which is almost similar to that of Gujarati Indians in US.
The frequency of the minor allele TNFα-308 (A) in the present cohort is low (0.02 in controls), which corroborates the findings of another study on North Indian population. [25] There is a wide variation in the incidence of the A allele in different populations across the world. The highest frequency (0.274) was recorded in a Caucasian population. [26] Data from the HapMap indicated frequency of the A allele in Europeans to be 0.217, whereas in the Japanese population it was 0.02.
The differences in the frequency of promoter polymorphisms of the cytokines are exciting, since they have functional significance and could partially explain interethnic variation found in other states such as autoimmune diseases.
In recent times, the interplay between inflammation and lipid metabolism has also been the focus of research aimed at understanding the mechanisms of atherogenesis. In the present study, no association of IL-6-174G/C genotypes with lipid levels was seen, which is in agreement with a study on healthy Mexican subjects. [27] In contrast, the Genetics of Lipid Lowering Drugs and Diet Network study, the GG and CG subjects showed higher fasting plasma TG (P = 0.025), VLDL (P = 0.04) and large VLDL (P 5 0.02) concentrations than did CC subjects. [28] No association of the TNFα-308 genotypes with any of the lipid parameters was seen in the present study, though in contrast the study on Mexican population indicated that high levels of TGs and TC, as well as low HDL-C levels were associated with GG genotype of the-308 TNFα polymorphism., [27] These contrasting results may be due to inherent difficulties in studying the interaction between inflammation and lipid metabolism in vivo due to anorexia triggered by the proinflammatory cytokines. [29]
No association with CAD risk was seen with the IL-6-174G > C SNP in our study, which is similar to the results of the Rotterdam Heart study, a meta-analysis [30] and the MONICA survey. [31] Our results are also in agreement with the Ludwigshafen Risk and Cardiovascular Health (LURIC) cohort study. [32] However, in contrast to our study and several others the ECTIM study implicated the-174C allele with over 1.34 times risk of MI. [33] Another study on Asian Indians observed the haplotype defined by this promoter SNPs to be associated with over 3.7 times risk of CAD (OR 3.676, 95% CI 1.68-8.05, P = 0.0017), [34] which is in contrast to our findings and also to that in a study on North Indians. [35]
In the present study, the TNFα-308G > A did not associate with increased CAD risk after accounting for the confounders. The findings of the present study are similar to that of Koch et al. where no association was seen with both IL-10 and TNFα promoter polymorphisms −863C > A and −308G > A, either alone or acting co-operatively with the risk of CAD/MI. [36] A study on the North Indian population also has found no association of this variant with risk of CAD. [37] Recent meta-analysis [38] has also concluded that "It is probable that carrying the-308A variant is associated with CAD risk in Caucasians, but not in Asians, Indians or Africans. Further studies are merited to assess the association in greater details, especially in Asians, Indians and Africans".
In summary, no association of the permissive promoter variants in the IL-6 gene and the TNFα gene were seen with increased CAD risk. Though, one of the limitations of the study is the sample size and larger studies are ongoing, these and other studies highlight the importance of doing population specific studies taking into account the different ethnicities.
 References References | |  |
| 1. | Ross R. Atherosclerosis - An inflammatory disease. N Engl J Med 1999;340:115-26. 
|
| 2. | Libby P, Ridker PM, Hansson GK, Leducq Transatlantic Network on Atherothrombosis. Inflammation in atherosclerosis: From pathophysiology to practice. J Am Coll Cardiol 2009;54:2129-38. 
|
| 3. | Charo IF, Ransohoff RM. The many roles of chemokines and chemokine receptors in inflammation. N Engl J Med 2006;354:610-21. 
|
| 4. | Viola A, Luster AD. Chemokines and their receptors: Drug targets in immunity and inflammation. Annu Rev Pharmacol Toxicol 2008;48:171-97. 
|
| 5. | Libby P, Theroux P. Pathophysiology of coronary artery disease. Circulation 2005;111:3481-8. 
|
| 6. | Sandhu MS, Waterworth DM, Debenham SL, Wheeler E, Papadakis K, Zhao JH, et al. LDL-cholesterol concentrations: A genome-wide association study. Lancet 2008;371:483-91. 
|
| 7. | Melzer D, Perry JR, Hernandez D, Corsi AM, Stevens K, Rafferty I, et al. A genome-wide association study identifies protein quantitative trait loci (pQTLs). PLoS Genet 2008;4:e1000072. 
|
| 8. | Wallace C, Newhouse SJ, Braund P, Zhang F, Tobin M, Falchi M, et al. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: Serum urate and dyslipidemia. Am J Hum Genet 2008;82:139-49. 
|
| 9. | Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet 2008;40:161-9. 
|
| 10. | Kroeger KM, Carville KS, Abraham LJ. The-308 tumor necrosis factor-alpha promoter polymorphism effects transcription. Mol Immunol 1997;34:391-9. 
|
| 11. | Rosenwasser LJ, Borish L. Promoter polymorphisms predisposing to the development of asthma and atopy. Clin Exp Allergy 1998;28 Suppl 5:13-5. 
|
| 12. | Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet 1997;24:1-8. 
|
| 13. | Licastro F, Chiapelli M, Caldarera CM, Caruso C, Lio D, Corder EH. Acute myocardial infarction and proinflammatory gene variants. Ann N Y Acad Sci 2007;1119:227-42. 
|
| 14. | Olomolaiye O, Wood NA, Bidwell JL. A novel NlaIII polymorphism in the human IL-6 promoter. Eur J Immunogenet 1998;25:267. 
|
| 15. | Fishman D, Faulds G, Jeffery R, Mohamed-Ali V, Yudkin JS, Humphries S, et al. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest 1998;102:1369-76. 
|
| 16. | Castell JV, Gómez-Lechón MJ, David M, Andus T, Geiger T, Trullenque R, et al. Interleukin-6 is the major regulator of acute phase protein synthesis in adult human hepatocytes. FEBS Lett 1989;242:237-9. 
|
| 17. | Wilson AG, Symons JA, McDowell TL, McDevitt HO, Duff GW. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc Natl Acad Sci U S A 1997;94:3195-9. 
|
| 18. | Louis E, Franchimont D, Piron A, Gevaert Y, Schaaf-Lafontaine N, Roland S, et al. Tumour necrosis factor (TNF) gene polymorphism influences TNF-alpha production in lipopolysaccharide (LPS)-stimulated whole blood cell culture in healthy humans. Clin Exp Immunol 1998;113:401-6. 
|
| 19. | Blake GJ, Ridker PM. Novel clinical markers of vascular wall inflammation. Circ Res 2001;89:763-71. 
|
| 20. | Nicola PJ, Maradit-Kremers H, Roger VL, Jacobsen SJ, Crowson CS, Ballman KV, et al. The risk of congestive heart failure in rheumatoid arthritis: A population-based study over 46 years. Arthritis Rheum 2005;52:412-20. 
|
| 21. | Ridker PM, Brown NJ, Vaughan DE, Harrison DG, Mehta JL. Established and emerging plasma biomarkers in the prediction of first atherothrombotic events. Circulation 2004;109:IV6-19. 
|
| 22. | Kilpinen S, Hulkkonen J, Wang XY, Hurme M. The promoter polymorphism of the interleukin-6 gene regulates interleukin-6 production in neonates but not in adults. Eur Cytokine Netw 2001;12:62-8. 
|
| 23. | Veres A, Prohászka Z, Kilpinen S, Singh M, Füst G, Hurme M. The promoter polymorphism of the IL-6 gene is associated with levels of antibodies to 60-kDa heat-shock proteins. Immunogenetics 2002;53:851-6. 
|
| 24. | Rauramaa R, Väisänen SB, Luong LA, Schmidt-Trücksäss A, Penttilä IM, Bouchard C, et al. Stromelysin-1 and interleukin-6 gene promoter polymorphisms are determinants of asymptomatic carotid artery atherosclerosis. Arterioscler Thromb Vasc Biol 2000;20:2657-62. 
|
| 25. | Gupta V, Sehajpal PK. Rapid detection of single nucleotide (-308) polymorphism in TNF-á promoter using ARMS-PCR. Curr Sci 2003;85:1521-3. 
|
| 26. | Perrey C, Pravica V, Sinnott PJ, Hutchinson IV. Genotyping for polymorphisms in interferon-gamma, interleukin-10, transforming growth factor-beta 1 and tumour necrosis factor-alpha genes: A technical report. Transpl Immunol 1998;6:193-7. 
|
| 27. | Parra-Rojas I, Ruíz-Madrigal B, Martínez-López E, Panduro A. Influence of the-308 TNF-alpha and-174 IL-6 polymorphisms on lipid profile in Mexican subjects. Hereditas 2006;143:167-72. 
|
| 28. | Shen J, Arnett DK, Pérez-Martínez P, Parnell LD, Lai CQ, Peacock JM, et al. The effect of IL6-174C/G polymorphism on postprandial triglyceride metabolism in the GOLDN studyboxs. J Lipid Res 2008;49:1839-45. 
|
| 29. | Grunfeld C, Zhao C, Fuller J, Pollack A, Moser A, Friedman J, et al. Endotoxin and cytokines induce expression of leptin, the ob gene product, in hamsters. J Clin Invest 1996;97:2152-7. 
|
| 30. | Sie MP, Sayed-Tabatabaei FA, Oei HH, Uitterlinden AG, Pols HA, Hofman A, et al. Interleukin 6-174 g/c promoter polymorphism and risk of coronary heart disease: Results from the rotterdam study and a meta-analysis. Arterioscler Thromb Vasc Biol 2006;26:212-7. 
|
| 31. | Lieb W, Pavlik R, Erdmann J, Mayer B, Holmer SR, Fischer M, et al. No association of interleukin-6 gene polymorphism (-174 G/C) with myocardial infarction or traditional cardiovascular risk factors. Int J Cardiol 2004;97:205-12. 
|
| 32. | Nauck M, Winkelmann BR, Hoffmann MM, Böhm BO, Wieland H, März W. The interleukin-6 G(-174) C promoter polymorphism in the LURIC cohort: No association with plasma interleukin-6, coronary artery disease, and myocardial infarction. J Mol Med (Berl) 2002;80:507-13. 
|
| 33. | Georges JL, Loukaci V, Poirier O, Evans A, Luc G, Arveiler D, et al. Interleukin-6 gene polymorphisms and susceptibility to myocardial infarction: The ECTIM study. Etude Cas-Témoin de l'Infarctus du Myocarde. J Mol Med (Berl) 2001;79:300-5. 
|
| 34. | Maitra A, Shanker J, Dash D, John S, Sannappa PR, Rao VS, et al. Polymorphisms in the IL6 gene in Asian Indian families with premature coronary artery disease - The Indian Atherosclerosis Research Study. Thromb Haemost 2008;99:944-50. 
|
| 35. | Banerjee I, Pandey U, Hasan OM, Parihar R, Tripathi V, Ganesh S. Association between inflammatory gene polymorphisms and coronary artery disease in an Indian population. J Thromb Thrombolysis 2009;27:88-94. 
|
| 36. | Koch W, Kastrati A, Böttiger C, Mehilli J, von Beckerath N, Schömig A. Interleukin-10 and tumor necrosis factor gene polymorphisms and risk of coronary artery disease and myocardial infarction. Atherosclerosis 2001;159:137-44. 
|
| 37. | Shanker J, Kakkar VV. Implications of genetic polymorphisms in inflammation-induced atherosclerosis. Open Cardiovasc Med J 2010;4:30-7. 
|
| 38. | Zhang HF, Xie SL, Wang JF, Chen YX, Wang Y, Huang TC. Tumor necrosis factor-alpha G-308A gene polymorphism and coronary heart disease susceptibility: An updated meta-analysis. Thromb Res 2011;127:400-5. 
|
[Figure 1]
[Table 1], [Table 2], [Table 3], [Table 4]
|






