|
 
 |
|
ORIGINAL ARTICLE |
|
|
|
| Year : 2014 | Volume
: 20
| Issue : 1 | Page : 32-36 |
| |
Pattern of chromosome involvement in childhood hyperdiploid pre-B-cell acute lymhoblastic leukemia cases from India
Lily S. Kerketta, Vundinti Baburao, Kanjaksha Ghosh
Department of Cytogenetics and Molecular Genetics, National Institute of Immunohaematology, KEM Hospital, Mumbai, Maharashtra, India
| Date of Web Publication | 19-May-2014 |
Correspondence Address:
Kanjaksha Ghosh
Department of Cytogenetics and Molecular Genetics, National Institute of Immunohaematology, 13th Floor, KEM Hospital, Parel, Mumbai, Maharashtra
India
 Source of Support: Indian Council of Medical Research, Conflict of Interest: None
DOI: 10.4103/0971-6866.132751

 Abstract Abstract | | |
Background: Hyperdiploid pre-B-cell acute lymhoblastic leukemia (pre-B-ALL) is a common form of childhood leukemia with very good prognosis with present day chemotherapy. However, the chromosomal composition of the hyperdiploidy has not been extensively studied and possible mechanism for this pathology remains so far conjectural.
Objective: To analyze the pattern of chromosome involvement in a cohort of childhood hyperdiploid pre-B-ALL from India and from this pattern to develop an understanding on the causation of such pathology. Whether such patients also carry translocations and FLT3 mutations in addition to their hyperdiploid karyotype.
Materials and Methods: One hundred and twenty-six childhood pre-B-ALL patients were studied. Bone marrow aspirate of these patients were evacuated for morphology, FAB classification, immunophenotyping and both conventional and molecular cytogenetics.
Results: Of 126 patients with pre-B-ALL (age 2-15 years), 90 patients with abnormal karyotype showed 50 with hyperdiploid karyotype (50/90 i.e. 55.5%). Chromosomes 9, 10, 14, 17, 18, 20 and 21 were more often involved in hyperdiploidy. Chromosome 21 duplication was present in 92% of the cases. Chromosomes 5, 15, 16, 17 and Y were less often involved (18-20%) in hyperdiploidy. About 44% of patients with hyperdiploidy had additional karyotypic abnormality of which TEL-AML1 was predominant (24%). Chromosome loss was rare and accounted for 20% of the cases only. We did not find any FLT3 mutation in our patients.
Conclusion: In this study, the pattern of chromosome involvement in hyperdiploid karyotype of ALL patients is same as other studies except some chromosomes like 1, 6, 11, 12, 19 and 22 have some more frequent involvement than other studies. This study also showed the occurrence of TEL/AML1 fusion is more (19.8%) than other reports from India.
Keywords: Abnormal mitosis, centrosome pathology, cytogenetics, hyperdiploidy, pre-B acute lymphoblastic leukemia, selective gain of chromosomes, uniparental disomy
How to cite this article:
Kerketta LS, Baburao V, Ghosh K. Pattern of chromosome involvement in childhood hyperdiploid pre-B-cell acute lymhoblastic leukemia cases from India. Indian J Hum Genet 2014;20:32-6 |
How to cite this URL:
Kerketta LS, Baburao V, Ghosh K. Pattern of chromosome involvement in childhood hyperdiploid pre-B-cell acute lymhoblastic leukemia cases from India. Indian J Hum Genet [serial online] 2014 [cited 2016 Aug 24];20:32-6. Available from: http://www.ijhg.com/text.asp?2014/20/1/32/132751 |
 Introduction Introduction | |  |
Hyperdiploidy is one of the common chromosomal abnormalities in the childhood pre-B acute lymphoblastic leukemia (pre-B-ALL) and involves 30% of patients with the disease. Hyperdiploid pre-B-ALL is one of the prognostically most favorable leukemia. [1] The cause of hyperdiploidy in this condition is not well known but several hypothesis exist. [2],[3],[4] One of the important question is whether all the chromosomes are randomly represented in this hyperdiploidy or there are some preferential chromosomes, which are present more often in multiple numbers. In hyperdiploidy certain chromosomes e.g. no. 4, 6, 10, 14, 17, 18, 20, 21 and X were found to be added frequently. [1] Certain chromosomes are also lost in this disease but there is paucity of data on the composition of chromosome loss. In this paper, we analyzed our data on the composition of hyperdiploid chromosomes in well-characterized cohort of childhood pre-B-ALL.
 Materials and Methods Materials and Methods | |  |
A total of 126 childhood pre-B-ALL patients presenting in various hospitals in Mumbai and referred to this institute for detailed work-up in the patient population based on complete blood count, peripheral smear examination and routine biochemistry including serum lactate dehydrogenase and uric acid levels. Bone marrow aspirate in all these patients were evaluated for morphology for FAB classification, immunophenotyping and both conventional and molecular cytogeneitc studies.
Immunophenotype was done in BD FACS Aria R (B-D) using monoclonal antibodies CD10, CD19, CD5, CD7, CD3, CD33, CD3, HLA-DR, peroxidase, CD34 and anti-kappa, anti-lambda Immunohematology antibodies. Gating was done with forward light scatter/laser scanning cytometry and CD45 positive areas. Low CD45 with CD19 was assessed. Pre-B-ALL was diagnosed as CD45 + , CD34 + , CD10 + , CD19 + , cytoplasmic IgG, CD33− and CD3− .
Cytogenetic study was done using standard technique with Giemsa banding. [5] Fluorescence in situ hybridization (FISH) studies were done in hyperdiploid cases with Bcr-Abl t (9;22), MLL-AF4 t (4;11), TEL-AML1 t (12;21) and E2A/PBX t (1;19) using Vysis probes and its technique described in the kit.
Giemsa banded karyotypes were analyzed by one of us (LK) and the chromosomes were counted and identified using standard International System for Cytogenetic nomenclature 2005 method. [6] FLT3 internal tandem duplication and tyrosine kinase domain (ITD and TKD) mutations were tested using standard techniques which were already applied in this laboratory. [7]
 Results Results | |  |
Out of 126 pre-B-ALL patients, 90 patients had abnormal karytoype/FISH studies, 24 had normal cytogenetic pattern and in 12 patients karyotyping failed and no FISH abnormality was seen with the probes used. 50/90 patients had hyperdiploid karyotypes and the present study involves these 50 patients with hyperdiploid Karyotype (male:female 33:17, age 2-15 years). Leucocyte count at presentation varied between 2 × 10 9 /L and 78 × 10 9 /L, hemoglobin 46-87 g/L and platelets 18 × 10 9 /L-67 × 10 9 /L. Bone marrow aspirate showed L1 morphology in 37 and L2 in 13 patients.
All 50 patients had different grades of hyperdiploidy. The distribution of different chromosomes in these hyperdiploid phenotypes are presented in [Figure 1] and [Table 1]. In this study, chromosome 21 had a major contribution for the formation of hyperdiploid karyotype.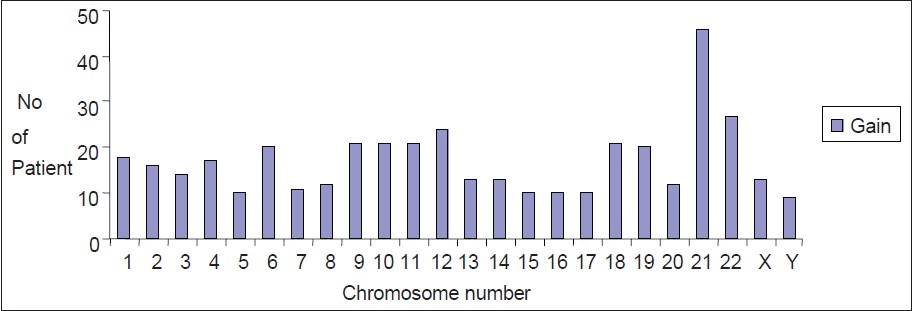 | Figure 1: Gain of chromosomes in childhood hyperdiploid pre-B cell acute lymphoblastic leukemia
Click here to view |
In 46 (92%) hyperdiploid patients, the number of an extra chromosome 21 varied between 1 and 5. There were only four patients (8%) where chromosome number 21 was not involved in hyperdiploidy and in 2 (4%) patient this chromosome was the only one which involved in hyperdiploidy. Other chromosomes were present in excess in 18-54% of the cases. Chromosomes 5, 15, 16, 17 and Y were less often involved (18-20%) in hyperdiploidy.
Individual chromosomal deletion was seen in hyperdiploid cases too. Chromosomes 2, 3, 4, 5, 6, 10, 21, X and Y showed no deletion. Chromosmes 1, 7, 8, 9, 11, 12, 14, 15, 22 showed deletion in 10/50 (20%) cases [Figure 2] and [Table 2]. Similarly, smaller percentage of patients showed deletion in other chromosomes. Twenty-two patients (22/50 i.e. 44%) had additional chromosomal abnormalities of which TEL-AML1 fusion was present in 12/50 (24%) of hyperdiploid patients. t (9;22) and t (4;11) were not seen in any of these patients though t (1;19) and t (12;20) were seen in 2% each [Table 3]. No FLT3 mutation (ITD/TKD) was observed in any of the patients.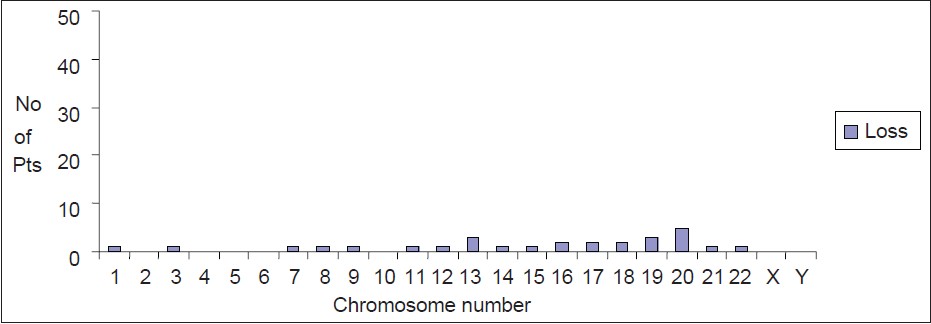 | Figure 2: Loss of chromosome in hyperdiploid childhood pre-B cell acute lymphoblastic leukemia patients
Click here to view |
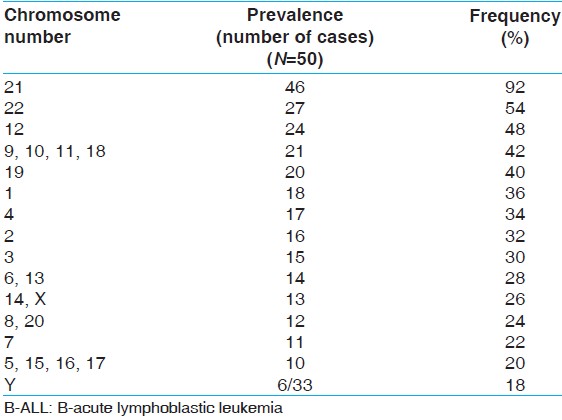 | Table 1: Prevalence of hyperdiploidy of different chromosomes in hyperdiploid childhood B-ALL cases
Click here to view |
 Discussion Discussion | |  |
Hyperdiploidy is a frequent phenomenon in childhood ALL and it is a good prognostic marker. [1],[2] In our series, 50/90 (55.5%) patients showing abnormal karyotype had hyperdiploidy and 50/126 (40%) cases of pediatric ALL showed hyperdiploidy, whether the karyotype failed or it was abnormal. This prevalence composes well with the prevalence described in the existing literatures. [2],[8] If we would have done DNA index study using flow cytometry (index >1.2) we could have obtained few more hyperdiploid patients where karyotype was failed. There are few studies who have evaluated the contribution of individual chromosome to hyperdiploidy with a view to understand (i) Why hyperdiploid chromosome pretends good prognosis (ii) what possible mechanism may under lie to produce hyperdiploid phenotype. Certain chromosomes e.g. 9, 10, 14, 17, 18, 20 and 21 are found to be more often involved in the hyperdiploid karyotype. [9],[10],[11] In our series chromosome 9, 10, 11, 12, 18, 21 and 22 were found to be frequently involved in 42 (92%) hyperdiploid cases [Table 1]. By far the most common chromosome involved in hyperdiploidy was chromosome 21 (92%) and 2 (4%) patients in our series had only this anomaly showing that this anomaly may be the primary one. 44% of our patients had additional chromosome abnormalities of which t (12;21) was found to be the most common (24%) one. Several studies have shown that t (12;21) that is TEL-AML1 fusion are present in prognostically favorable group of ALL patients. Prevalence of this condition in an unselected group of ALL patients in India have shown between 4% and 9% of the cases depending on the technique used to detect this abnormality. [12],[13],[14] In our series of 126 patients 19.8% that is, 25/126 patients showed this abnormality. Hence TEL-AML1 fusion in both hyperdiploid 12/50 (24%) and nonhyperdiploid 22/74 (29%) are not statistically different (y2 test, P > 0.05).
Though chromosome 21 was present in excess number in 92% of hyperdiploid karyotype, in 8% of the patients this chromosome was not involved, in fact none of these 8% patient had any chromosomal deletion too. Deletion of chromosomes seems to be poorly tolerated in ALL patients. Only 10/50 (20%) of the ALL patient showed one or more deletions. Single chromosome deletion was seen in 60% of these cases. In 2/50 (4%) cases chromosome 17 was deleted and this chromosome is well-known for the gene P53 whose importance as gate keeper of the genome is very well-known. Hence, involvement of chromosome 21 in hyperdiploid pediatric ALL seems to be the primary pathology. We could not do refined studies with extended molecular probes for chromosome 21 to see if the 4 of our patients where chromosome 21 involvement was seen, did any additional chromosome 21 segment hidden within the other chromosomes. About 44% of our patients had additional chromosomal abnormalities where t (12;21) was the most common one. However, none of the four patients those who did not have additional chromosome no. 21 had any (12;21) translocation.
One of the root questions in this subtype of ALL is how this hyperdiploidy arise? (i) Is it by mitotic events when a haploid cell is produced and subsequent mitosis and loss of chromosome leads to hyperdiploidy [3],[15] or a single mitotic event producing hyperdiploid (tetraploid) karyotype then loss of chromosomes in subsequent cell division cause the detectable karyotypic change. [4],[16] Our study suggest none of the two mechanism can explain infrequent loss of chromosome with highly gain in chromosome number 21. Sometimes, the gain in individual chromosome can vary from +1 to +5. Hence, doubling the haploid set of chromosomes cannot produce this.
Endoreduplication of chromosomes due to silencing of some of the cell cycle controlling genes cause natural polyploidy in cells like megakaryocytes. It may be possible that similar such mechanism coupled with weakness of the mitotic spindle and which cause loss of chromosome in subsequent mitosis. This loss mainly involves the additional chromosomes over and above the diploid sets. However, this pathology also fails to explain why we have a high rate of translocation events in addition to hyperdiploidy in childhood hyperdiploid pre-B-ALL. Unless we assume that some of these mooring less extra chromosomes interact with other chromosome to produce the translocation or partial deletion events.
To explain why hyperdiploid karyotype in pediatric B-ALL is associated with good prognosis, it has been shown that these cells are extremely sensitive to methotrexate and one of the folate carrier protein e.g. reduced folate carrier (RFC) gene is encoded in chromosome 21. Hence multiple copies of chromosome 21 probably lead to high amount of RFC in the cell leading to higher methotrexate concentration in the cell. [17] Present study shows that in addition to involvement of chromosome 21 as hyperdiploidy a large proportion (24%) of these cases also have additional chromosome pathology e.g. TEL-AML1 fusion, which may contributes to improve prognosis in these cases.
Several authors have shown high FLT3 mutation in hyperdiploid ALL patients however in our series we could not demonstrate either ITD or TKD mutation of FLT3 gene. [18],[19],[20] Whether this finding is true for Indian pediatric ALL patients? Though, the present report does not conclusively provide the mechanism of hyperdiploidy in ALL except as a conjecture. It definitely shows that existing hypothesis of hyperdiploidy are inadequate to explain the phenomenon in its entirety.
 Acknowledgements Acknowledgements | |  |
Financial Assistance from Indian Council of Medical Research is gratefully acknowledged.
 References References | |  |
| 1. | Paulsson K, Johansson B. High hyperdiploid childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer 2009;48:637-60. 
|
| 2. | Onodera N, McCabe NR, Rubin CM. Formation of a hyperdiploid karyotype in childhood acute lymphoblastic leukemia. Blood 1992;80:203-8. 
|
| 3. | Onodera N, McCabe NR, Nachman JB, Johnson FL, Le Beau MM, Rowley JD, et al. Hyperdiploidy arising from near-haploidy in childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer 1992;4:331-6. 
|
| 4. | Paulsson K, Mörse H, Fioretos T, Behrendtz M, Strömbeck B, Johansson B. Evidence for a single-step mechanism in the origin of hyperdiploid childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer 2005;44:113-22. 
|
| 5. | Seabright M. A rapid banding technique for human chromosomes. Lancet 1971;2:971-2. 
[PUBMED] |
| 6. | Heerema NA, Sather HN, Sensel MG, Zhang T, Hutchinson RJ, Nachman JB, et al. Prognostic impact of trisomies of chromosomes 10, 17, and 5 among children with acute lymphoblastic leukemia and high hyperdiploidy (and gt; 50 chromosomes). J Clin Oncol 2000;18:1876-87. 
|
| 7. | Kumar S, DeCamillo D, Bhambhani K, Cushing B, Thomas R, Mohamed AN, et al. Children with hyperdiploid but not triple trisomy(+4, +10, +7 acute lymphoblastic leukemia have an increased incidence of extramedullary relapse on current therapies: A single institution experience. Am J Hematol 2008;83:34-40. 
|
| 8. | Settin A, Al Haggar M, Al Dosoky T, Al Baz R, Abdelrazik N, Fouda M, et al. Prognostic cytogenetic markers in childhood acute lymphoblastic leukemia. Indian J Pediatr 2007;74:255-63. 
|
| 9. | Paulsson K, Panagopoulos I, Knuutila S, Jee KJ, Garwicz S, Fioretos T, et al. Formation of trisomies and their parental origin in hyperdiploid childhood acute lymphoblastic leukemia. Blood 2003;102:3010-5. 
|
| 10. | Hamouda F, El-Sissy AH, Radwan AK, Hussein H, Gadallah FH, Al-Sharkawy N, et al. Correlation of karyotype and immunophenotype in childhood acute lymphoblastic leukemia; experience at the National Cancer Institute, Cairo University, Egypt. J Egypt Natl Canc Inst 2007;19:87-95. 
|
| 11. | Liang DC, Shih LY, Yang CP, Hung IJ, Liu HC, Jaing TH, et al. Frequencies of ETV6-RUNX1 fusion and hyperdiploidy in pediatric acute lymphoblastic leukemia are lower in far east than west. Pediatr Blood Cancer 2010;55:430-3. 
|
| 12. | Sudhakar N, Rajalekshmy KR, Rajkumar T, Nancy KN. RT-PCR and real-time PCR analysis of E2A-PBX1, TEL-AML1, mBCR-ABL and MLL-AF4 fusion gene transcripts in de novo B-lineage acute lymphoblastic leukaemia patients in south India. J Genet 2011;90:349-53. 
|
| 13. | Sazawal S, Bhatia K, Gutierrez MI, Saxena R, Arya LS, Bhargava M. Paucity of TEL-AML 1 translocation, by multiplex RT-PCR, in B-lineage acute lymphoblastic leukemia (ALL) in Indian patients. Am J Hematol 2004;76:80-2. 
|
| 14. | Inamdar N, Kumar SA, Banavali SD, Advani S, Magrath I, Bhatia K. Comparative incidence of the rearrangements of TEL/AML1 and ALL1 genes in pediatric precursor B acute lymphoblastic leukemias in India. Int J Oncol 1998;13:1319-22. 
|
| 15. | Panzer-Grümayer ER, Fasching K, Panzer S, Hettinger K, Schmitt K, Stöckler-Ipsiroglu S, et al. Nondisjunction of chromosomes leading to hyperdiploid childhood B-cell precursor acute lymphoblastic leukemia is an early event during leukemogenesis Blood 2002;100:347-9. 
|
| 16. | Attarbaschi A, Mann G, König M, Steiner M, Dworzak MN, Gadner H, et al. Near-tetraploidy in childhood B-cell precursor acute lymphoblastic leukemia is a highly specific feature of ETV6/RUNX1-positive leukemic cases. Genes Chromosomes Cancer 2006;45:608-11. 
|
| 17. | Zhang L, Taub JW, Williamson M, Wong SC, Hukku B, Pullen J, et al. Reduced folate carrier gene expression in childhood acute lymphoblastic leukemia: Relationship to immunophenotype and ploidy. Clin Cancer Res 1998;4:2169-77. 
|
| 18. | Armstrong SA, Mabon ME, Silverman LB, Li A, Gribben JG, Fox EA, et al. FLT3 mutations in childhood acute lymphoblastic leukemia Blood 2004;103:3544-6. 
|
| 19. | Braoudaki M, Karpusas M, Katsibardi K, Papathanassiou Ch, Karamolegou K, Tzortzatou-Stathopoulou F. Frequency of FLT3 mutations in childhood acute lymphoblastic leukemia. Med Oncol 2009;26:460-2. 
|
| 20. | Chang P, Kang M, Xiao A, Chang J, Feusner J, Buffler P, et al. FLT3 mutation incidence and timing of origin in a population case series of pediatric leukemia. BMC Cancer 2010;10:513. 
|
[Figure 1], [Figure 2]
[Table 1], [Table 2], [Table 3]
|






