|
 
 |
|
ORIGINAL ARTICLE |
|
|
|
| Year : 2014 | Volume
: 20
| Issue : 1 | Page : 37-42 |
| |
Association study of the ABCC8 gene variants with type 2 diabetes in south Indians
Radha Venkatesan1, Dhanasekaran Bodhini1, Nagarajan Narayani1, Viswanathan Mohan2
1 Department of Molecular Genetics, World Health Organization Collaborating Centre for Non Communicable Diseases Prevention and Control, International Diabetes Federation Centre for Education, Gopalapuram, Chennai, Tamil Nadu, India
2 Diabteology, Madras Diabetes Research Foundation and Dr. Mohan's Diabetes Specialities Centre, World Health Organization Collaborating Centre for Non Communicable Diseases Prevention and Control, International Diabetes Federation Centre for Education, Gopalapuram, Chennai, Tamil Nadu, India
| Date of Web Publication | 19-May-2014 |
Correspondence Address:
Radha Venkatesan
Department of Molecular Genetics, Madras Diabetes Research Foundation, 4, Conransmith Road, Gopalapuram, Chennai 600 086, Tamil Nadu
India
 Source of Support: This work was funded by Indian Council for Medical
Research (ICMR),, Conflict of Interest: None
DOI: 10.4103/0971-6866.132752

 Abstract Abstract | | |
Background: The ABCC8 gene which encodes the sulfonylurea receptor plays a major role in insulin secretion and is a potential candidate for type 2 diabetes. The -3c → t (rs1799854) and Thr759Thr (C → T, rs1801261) single nucleotide polymorphisms (SNPs) of the ABCC8 gene have been associated with type 2 diabetes in many populations. The present study was designed to investigate the association of these two SNPs in an Asian Indian population from south India.
Materials and Methods: A total of 1,300 subjects, 663 normal glucose tolerant (NGT) and 637 type 2 diabetic subjects were randomly selected from the Chennai Urban Rural Epidemiology Study (CURES). The -3c → t and Thr759Thr were genotyped in these subjects using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) and a few variants were confirmed by direct sequencing.
Results: The frequency of the 't' allele of the -3c → t SNP was found to be 0.27 in NGT and 0.29 in type 2 diabetic subjects (P = 0.44). There was no significant difference in the genotypic frequency between the NGT and type 2 diabetic group (P = 0.18). Neither the genotypic frequency nor the allele frequency of the Thr759Thr polymorphism was found to differ significantly between the NGT and type 2 diabetic groups.
Conclusion: The -3c → t and the Thr759Thr polymorphisms of the ABCC8 gene were not associated with type 2 diabetes in this study. However, an effect of these genetic variants on specific unidentified sub groups of type 2 diabetes cannot be excluded.
Keywords: ABCC8 gene , south Indians, sulfonylurea receptor, type 2 diabetes
How to cite this article:
Venkatesan R, Bodhini D, Narayani N, Mohan V. Association study of the ABCC8 gene variants with type 2 diabetes in south Indians. Indian J Hum Genet 2014;20:37-42 |
How to cite this URL:
Venkatesan R, Bodhini D, Narayani N, Mohan V. Association study of the ABCC8 gene variants with type 2 diabetes in south Indians. Indian J Hum Genet [serial online] 2014 [cited 2016 Aug 24];20:37-42. Available from: http://www.ijhg.com/text.asp?2014/20/1/37/132752 |
FNx01Equal contribution
 Introduction Introduction | |  |
The sulfonylurea receptor (SUR) is a subunit of the ATP-sensitive potassium channel in the pancreatic beta cell. This protein complex is involved in regulation of insulin secretion from the beta cell in response to glucose. [1] The gene encoding the sulfonylurea receptor, ABCC8 gene (located at chromosome 11p 15) is a candidate gene for type 2 diabetes as one of the major pathophysiological defects of type 2 diabetes is impaired insulin secretion. The -3c → t (rs1799854) and the Thr759Thr (rs1801261) variants of the ABCC8 gene have been widely studied in association with type 2 diabetes but the results have been inconsistent. [2],[3],[4],[5],[6],[7],[8],[9] A majority of these studies have been on white populations and there is very limited data in Asian Indians [10],[11] who have greater insulin resistance, increased susceptibility to type 2 diabetes, and a strong genetic background compared to Europeans. [12] India currently has 62.4 million people with diabetes [13] and it would be pertinent to study the role of these two polymorphisms in association with diabetes in this population. Hence, the present study was designed to investigate the association of -3c → t and the Thr759Thr polymorphims of the ABCC8 gene with type 2 diabetes in an Asian Indian population from south India.
 Materials and Methods Materials and Methods | |  |
Ethical approval
The present study was planned in agreement with the World Medical Association Declaration of Helsinki (2001). Informed consent was obtained from all the subjects who participated in this study and the study was approved by the institutional ethical committee.
Subjects
A total of 1,300 unrelated subjects were chosen from the Chennai Urban Rural Epidemiology Study (CURES). The methodology of the study has been published elsewhere. [14] In Phase 1 of CURES; 26,001 subjects were recruited based on a systematic random sampling technique. Self-reported diabetic subjects were classified as 'known diabetic subjects'. In Phase 2 of CURES, all known diabetic subjects (n = 1,529) were invited to our center for detailed studies of whom 1,382 responded. In Phase 3 of CURES, every 10 th individual from Phase 1 (n = 2,600) was invited to undergo oral glucose tolerance test (OGTT) using 75 g oral glucose load (dissolved in 250 ml of water). Those who had 2 h plasma glucose value ≥ 11.1 mmol/l (200 mg/dl) (based on World Health Organization (WHO) consulting group criteria) were labeled as 'newly detected diabetic subjects' (n = 222). Subjects who had fasting plasma glucose < 5.6 mmol/l (100 mg/dl) and 2 h plasma glucose value ≤ 7.8 mmol/l (140 mg/dl) were categorized as normal glucose tolerance [15] (NGT, n = 1736). The total number of diabetic subjects in the CURES study population is 1,604 (1,382 known diabetic subjects + 222 newly detected diabetic subjects). From these 1,604 diabetic subjects, 637 subjects, and from the 1,736 NGT subjects, 663 subjects were randomly selected for the present study.
Biochemical measurements
Anthropometric measurements including weight and height were obtained using standardized techniques. The body mass index (BMI) was calculated as the weight in kilograms divided by the square of height in meters. Biochemical analyses were carried out on a Hitachi-912 Autoanalyzer (Hitachi, Mannheim, Germany) using commercial kits (Roche Diagnostics, Mannheim, Germany). Fasting plasma glucose was estimated using glucose oxidase-peroxidase method. Serum cholesterol was estimated using cholesterol oxidase-phenol 4-amino antipyrene peroxidase (CHOD-PAP) method. Serum triglyceride was estimated using glycerol phosphatase oxidase-PAP (GPO-PAP) method. High-density lipoprotein (HDL) cholesterol was estimated using polyethylene glycol-pretreated enzymes method and low-density lipoprotein (LDL) cholesterol was calculated using Friedewald formula. Glycated hemoglobin (HbA1C) was estimated by high performance liquid chromatography using the Variant machine (Bio-Rad, Hercules, USA). Serum insulin concentration was estimated using Dako kits (Dako, Glostrup, Denmark).
Genotyping
DNA was isolated from whole blood using the phenol-chloroform method. The -3c → t (rs1799854) and the Thr759Thr (C → T, rs1801261) polymorphisms were genotyped by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) using the following primers: forward, 5'- GAG CCA GAG GAG GAT GTT GA-3', reverse 5'- GGC TAG AAG GAG CGA GGA CT-3' and forward 5'- TAA AGG CAT GCT CAT GTT GG-3', reverse 5'- AAT GTT CCC AGG ACG CAG TA-3' (Sigma, Bangalore, India), respectively. A final reaction volume of 15 μL of polymerase chain reaction contained 100 ng genomic DNA, 5 pmol of each primer, polymerase chain reaction buffer with 1 mmol/L of MgCl 2 , 100 μmol/L of each deoxynucleotide triphospate (dNTP), and 0.5 U of Taq polymerase (Gibco, Life Technologies, New York, NY). Polymerase chain reaction was carried out on a Peltier Thermal Cycler (PTC-200, MJ Research, Waltham, MA) under the following conditions: 95°C for 5 min, followed by 34 cycles of 95°C for 30 s, 59°C (for -3c → t)/54°C (for Thr759Thr) for 30 s, 72°C for 30 s, and a final extension of 72°C for 9 min. RFLP was carried out using PstI enzyme for the -3c → t polymorphism and BsiE1 enzyme for the Thr759Thr polymorphism. The resulting products were electrophoresed on a 3% agarose gel. To assure that the genotyping was of sufficient quality, random duplication in about 20% of the samples was performed by a technician who was blinded to the phenotype. There was 99% concordance in the genotyping.
Statistical analysis
Statistical Package for Social Sciences (SPSS) Windows, version 10.0, was used for statistical analysis. One-way analysis of variance (ANOVA) was used to compare groups for continuous variables. Data for continuous variables were expressed as mean ± SD. Chi-square test was used to compare the proportions of genotypes or alleles. P values < 0.05 were considered statistically significant.
 Results Subject characteristics Results Subject characteristics | |  |
The -3c/t and the Thr759Thr polymorphisms were genotyped in 663 NGT subjects and 637 type 2 diabetic subjects. The diabetic subjects were older compared to the NGT subjects (49 ± 10 and 46 ± 11 years, respectively) [Table 1]. A comparison between diabetic and NGT subjects showed that the age- and sex-adjusted BMI, waist circumference, fasting plasma glucose, glycated hemoglobin, fasting serum insulin, total cholesterol, serum triglycerides, and LDL cholesterol were all significantly higher in the type 2 diabetic subjects (P < 0.001).
-3c/t polymorphism
[Figure 1] shows the ethidium bromide stained agarose gel photograph of the cc, ct, and tt genotypes of the -3c/t polymorphism. [Table 2] shows the genotypic and allelic frequency of the -3c/t polymorphism. The genotypic distribution was in Hardy-Weinberg equilibrium. The frequency of the 't' allele of the -3c/t polymorphism was found to be 27% in the NGT subjects and 29% in the type 2 diabetic subjects and the difference was not statistically significant (odds ratio (OR): 1.07, 95% confidence interval (CI): 0.90-1.27, P = 0.44). The genotypic distribution was not significantly different between the NGT and type 2 diabetic groups (P = 0.18). Logistic regression analysis taking diabetes status as the dependent variable and genotypes as the independent variable showed that the ct and tt genotypes were not associated with diabetes. The odds ratios for the ct and tt genotypes as compared to the cc genotype was found to be 1.22 (95% CI: 0.97-1.54, P = 0.09) and 0.95 (0.64-1.43, P = 0.89), respectively.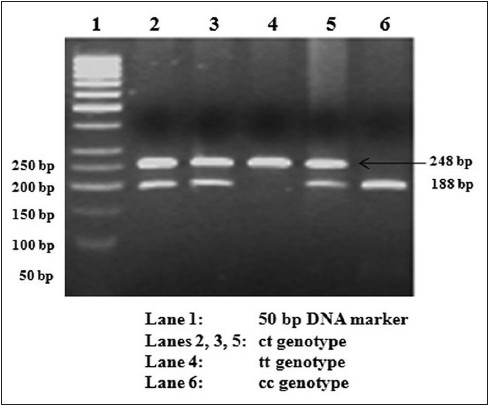 | Figure 1: Restriction fragment length polymorphism (RFLP) gel photograph of rs1799854 (-3c → t) polymorphism in ABCC8 gene
Click here to view |
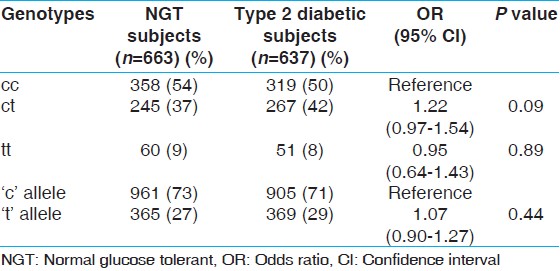 | Table 2: Genotype and allele frequency of the -3c?#168;t polymorphism studied in the ABCC8 gene
Click here to view |
When the clinical and biochemical parameters of the NGT and type 2 diabetic groups stratified according to the cc, ct, and tt genotypes were compared, none of the parameters showed any significant difference in the NGT or type 2 diabetic groups. Though the NGT subjects with the ct genotype had higher fasting serum insulin levels (mean ± standard deviation (SD): 9.59 ± 5.93 μIU/ml) compared to the cc genotype (9.16 ± 6.18 μIU/ml), the difference was not statistically significant.
Thr759Thr polymorphism
[Figure 2] shows the RFLP gel photograph of the Thr759Thr polymorphism. The genotype and allele frequency of this polymorphism has been presented in [Table 3]. The frequency of the heterozygous CT genotype of the Thr759Thr polymorphism was found to be similar in the NGT and type 2 diabetic subjects (0.5%). The homozygous variant TT genotype was not found in the study population. The frequency of the 'T' allele of Thr759Thr was only 0.2% in both the NGT and type 2 diabetic groups [Table 3]. A comparison of the biochemical parameters in the NGT and type 2 diabetic groups was not performed because of the very low frequency of the CT genotype.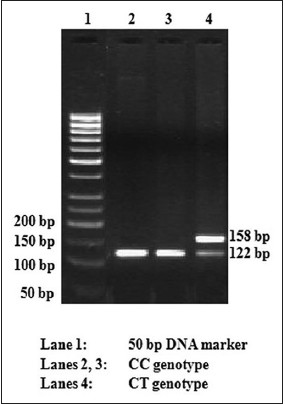 | Figure 2: RFLP gel photograph of rs1801261 (Thr759Thr, C → T) polymorphism in ABCC8 gene
Click here to view |
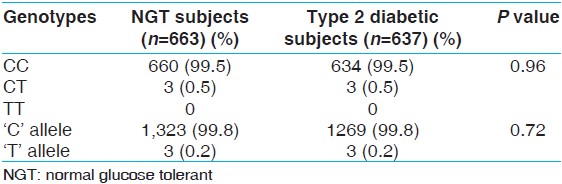 | Table 3: Genotype and allele frequency of the Thr759Thr polymorphism studied in the ABCC8 gene
Click here to view |
LD estimation between -3c→ t and the Thr759Thr single nucleotide polymorphisms (SNPs) and haplotype analysis
LD estimation between the -3c → t and the Thr759Thr SNPs showed that the pairwise LD between these SNPs was not high (r 2 = 0.001). In order to perform haplotype based analyses, two locus haplotypes were constructed and analysis was restricted to those haplotypes which have frequency of at least 0.01 in either cases or controls. Only two haplotypes, cC and tC were found to have a frequency >1%. There was no significant difference in the proportion of these haplotypes between NGT and type 2 diabetic groups (P = 0.58 and 0.73, respectively) [Table 4].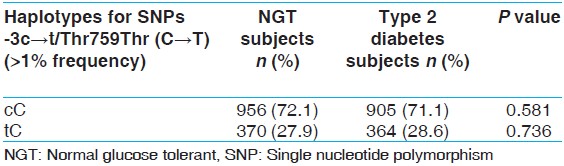 | Table 4: Comparison of frequencies of haplotypes in NGT and type 2 diabetic subjects
Click here to view |
 Discussion Discussion | |  |
The -3c → t (positioned in IVS15, 3bp ahead of exon 16) polymorphism and the Thr759Thr (AC→ AC) silent polymorphism (positioned in exon 18) were initially found to be associated with type 2 diabetes in Caucasians. [2] Subsequent studies on these two variants have been positive in Danish [3] and French Caucasian diabetic individuals, [4] and the -3c → t variant alone was associated in Dutch Caucasian diabetic subjects. [5] These two polymorphisms have also been studied in relation to insulin secretion in various populations and the results have been inconsistent. [6],[7],[8],[9] Two studies from north India did not find any association of the -3c → t (rs1799854) variant with type 2 diabetes. [10],[11] A number of factors, environmental, statistical, and genetic could underlie the variation in results observed in different populations. Non-replication of the association of specific locus alleles between populations of different ethnic backgrounds could be simply due to statistical bias from factors such as underlying population stratification. Alternatively, association with the same variant allele may be undetectable or different between ethnic groups because of differences in relative risks or pleiotropic genotype effects on phenotypic traits. Hence, replication studies within the same ethnic group are a more robust choice especially in populations such as Indians, who have greater insulin resistance and increased susceptibility to type 2 diabetes. [12] In this respect, this study testing the association of the two variants in the ABCC8 gene with type 2 diabetes in south Indian population gains significance.
The frequency of the 't' allele of the -3c → t polymorphism in this south Indian population (NGT - 0.27, type 2-0.29, P = 0.44) was much lesser as compared to the other populations such as the Finnish [7] (NGT - 0.43, type 2-0.60, P = 0.009), Danish Caucasians [3] (NGT - 0.44, type 2-0.45, P not significant), and Dutch Caucasians [5] (NGT - 0.41, type 2-0.48, P = 0.01); and did not show any significant difference between the NGT and type 2 diabetic groups. The frequency of the 'T' allele of the Thr759Thr polymorphism (0.2%) in our population appears to be the lowest among all populations studied so far. The fact that the homozygous TT genotype was not found in our population is comparable to the study on Dutch Caucasians [5] where the frequency of the TT genotype has been found to be <0.01.
In this study, both the variants were not found to be associated with type 2 diabetes. Similar to the present study, a number of other studies have also ruled out a major impact of the exon 16 and 18 variants in the ABCC8 gene on type 2 diabetes. A study which tested the association of 31 common SNPs with type 2 diabetes and its related traits in north Indian sibling pairs ruled out an association of the -3c → t polymorphism with type 2 diabetes, fasting glucose, and fasting insulin. [10] Another study on the association of 19 genes involved in pancreatic beta cell development and function with type 2 diabetes in north Indians [11] included several variants from the ABCC8 gene. The -3c → t polymorphism failed to show significant association with type 2 diabetes after correcting for multiple comparisons in that study. In a large study of Caucasians from the UK Prospective Diabetes Study (UKPDS) no association was seen between either exon 16 or 18 variant and diabetes. [16] The same was observed in independent populations from Netherlands, [17] Dutch Breda cohort, [18] and Japanese population. [19] A meta-analysis of reported association studies in Caucasian populations involving more than 7,000 subjects for the exon 16 and 18 variants did not support a substantial role of these two variants in the etiology of common type 2 diabetes. [20] The meta-analysis also showed that studies of the ABCC8 variants that were published first or had smaller sample sizes showed stronger associations. Publication bias might have affected the findings for the ABCC8 exon 16 and 18 variants. There is also not enough data to support a functional role of either of the two ABCC8 variants.
In conclusion, the -3c → t (rs1799854) and the Thr759Thr (rs1801261) polymorphisms of the ABCC8 gene were not associated with type 2 diabetes in this urban south Indian population. However, an effect of these genetic variants on specific unidentified subgroups of type 2 diabetes cannot be excluded.
 Acknowledgements Acknowledgements | |  |
We thank the ICMR for nominating MDRF as an ICMR-Advanced Centre for Genomics of type 2 diabetes. This is the 112 th publication from the CURES study (CURES 112).
 References References | |  |
| 1. | Miki T, Nagashima K, Seino S. The structure and function of the ATP-sensitive K+channel in insulin-secreting pancreatic beta-cells. J Mol Endocrinol 1999;22:113-23. 
|
| 2. | Inoue H, Ferrer J, Welling CM, Elbein SC, Hoffman M, Mayorga R, et al. Sequence variants in the sulfonylurea receptor (SUR) gene are associated with NIDDM in caucasians. Diabetes 1996;45:825-31. 
|
| 3. | Hansen T, Echwald SM, Hansen L, Møller AM, Almind K, Clausen JO, et al. Decreased tolbutamide-stimulated insulin secretion in healthy subjects with sequence variants in the high-affinity sulfonylurea receptor gene. Diabetes 1998;47:598-605. 
|
| 4. | Hani EH, Clément K, Velho G, Vionnet N, Hager J, Philippi A, et al. Genetic studies of the sulfonylurea receptor gene locus in NIDDM and in morbid obesity among French caucasians. Diabetes 1997;46:688-94. 
|
| 5. | Hart LM, de Knijff P, Dekker JM, Stolk RP, Nijpels G, van der does FE, et al. Variants in the sulphonylurea receptor gene: Association of the exon 16-3t variant with Type II diabetes mellitus in Dutch Caucasians. Diabetologia 1999;42:617-20. 
|
| 6. | Hart LM, Dekker JM, Van Haeften TW, Ruige JB, Stehouwer CD, Erkelens DW, et al. Reduced second phase insulin secretion in carriers of a sulphonylurea receptor gene variant associating with Type II diabetes mellitus. Diabetologia 2000;43:515-9. 
|
| 7. | Rissanen J, Markkanen A, Kärkkäinen P, Pihlajamäki J, Kekäläinen P, Mykkänen L, et al. Sulfonylurea receptor 1 gene variants are associated with gestational diabetes and type 2 diabetes but not with altered secretion of insulin. Diabetes Care 2000;23:70-3. 
|
| 8. | Weisnagel SJ, Rankinen T, Nadeau A, Rao DC, Chagnon YC, Pérusse L, et al. Decreased fasting and oral glucose stimulated C-peptide in nondiabetic subjects with sequence variants in the sulfonylurea receptor 1 gene. Diabetes 2001;50:697-702. 
|
| 9. | Reis AF, Hani EH, Beressi N, Robert JJ, Bresson JL, Froguel P, et al. Allelic variation in exon 18 of the sulfonylurea receptor 1 (SUR1) gene, insulin secretion and insulin sensitivity in nondiabetic relatives of type 2 diabetic subjects. Diabetes Metab 2002;28:209-15. 
|
| 10. | Gupta V, Vinay DG, Rafiq S, Kranthikumar MV, Janipalli CS, Giambartolomei C, et al. Indian Migration Study Group. Association analysis of 31 common polymorphisms with type 2 diabetes and its related traits in Indian sib pairs. Diabetologia 2012;55:349-57. 
|
| 11. | Chavali S, Mahajan A, Tabassum R, Dwivedi OP, Chauhan G, Ghosh S, et al. Association of variants in genes involved in pancreatic β-cell development and function withtype 2 diabetes in North Indians. J Hum Genet 2011;56:695-700. 
|
| 12. | Radha V, Mohan V. Genetic predisposition to type 2 diabetes among Asian Indians. Indian J Med Res 2007;125:259-74. 
[PUBMED]  |
| 13. | Anjana RM, Pradeepa R, Deepa M, Datta M, Sudha V, Unnikrishnan R, et al. ICMR-INDIAB Collaborative Study Group. Prevalence of diabetes and prediabetes (impaired fasting glucose and/or impaired glucose tolerance) in urban and rural India: Phase I results of the Indian Council of Medical Research-INdia DIABetes (ICMR-INDIAB) study. Diabetologia 2011;54:3022-7. 
|
| 14. | Deepa M, Pradeepa R, Rema M, Anjana M, Deepa R, Shanthirani S, et al. The Chennai Urban Rural Epidemiology Study (CURES)--study design and methodology (urban component) (CURES-1). J Assoc Physicians India 2003;51:863-70. 
|
| 15. | Alberti KG, Zimmet PZ. Definition diagnosis and classification of diabetes mellitus and its complications. Part 1: Diagnosis and classification of diabetes mellitus, provisional report of a WHO Consultation. Diabet Med 1998;15:539-53. 
[PUBMED] |
| 16. | Gloyn AL, Hashim Y, Ashcroft SJ, Ashfield R, Wiltshire S, Turner RC; UK Prospective Diabetes Study (UKPDS 53). Association studies of variants in promoter and coding regions of beta-cell ATP-sensitive K-channel genes SUR1 and Kir6.2 with Type 2 diabetes mellitus (UKPDS 53). Diabet Med 2001;18:206-12. 
|
| 17. | 't Hart LM, van Haeften TW, Dekker JM, Bot M, Heine RJ, Maassen JA. Variations in insulin secretion in carriers of the E23K variant in the Kir6·2 subunit of the ATP-sensitive K+channel in the beta-cell. Diabetes 2002;51:3135-8. 
|
| 18. | van Tilburg JH, Rozeman LB, Van Someren H, Rigters-Aris CA, Freriks JP, Pearson PL, et al. The exon 16-3t variant of the sulphonylurea receptor gene is not a risk factor for Type II diabetes mellitus in the Dutch Breda cohort. Diabetologia 2000;43:681-2. 
[PUBMED] |
| 19. | Yokoi N, Kanamori M, Horikawa Y, Takeda J, Sanke T, Furuta H, et al. Association studies of variants in the genes involved in pancreatic beta-cell function in type 2 diabetes in Japanese subjects. Diabetes 2006;55:2379-86. 
|
| 20. | van Dam RM, Hoebee B, Seidell JC, Schaap MM, de Bruin TW, Feskens EJ. Common variants in the ATP-sensitive K+channel genes KCNJ11 (Kir6.2) and ABCC8 (SUR1) in relation to glucose intolerance: Population based studies and meta-analyses. Diabet Med 2005;22:590-8. 
|
[Figure 1], [Figure 2]
[Table 1], [Table 2], [Table 3], [Table 4]
|






