|
 
 |
|
ORIGINAL ARTICLE |
|
|
|
| Year : 2014 | Volume
: 20
| Issue : 2 | Page : 175-184 |
| |
Prevalence of genetic variants associated with cardiovascular disease risk and drug response in the Southern Indian population of Kerala
Lakshmi Mahadevan1, Ancy Yesudas1, PK Sajesh1, S Revu1, Prasanna Kumar1, Devi Santhosh1, Sam Santhosh1, JM Sashikumar2, VK Gopalakrishnan3, Joji Boben4, Changanamkandath Rajesh1
1 MedGenome (Division of Molecular Diagnostics), SciGenom Labs Pvt. Ltd, CSEZ, Kakkanad, Cochin, Kerala, India
2 Department of Biotechnology, Karpagam University, Coimbatore, Tamil Nadu, India
3 Department of Biochemistry, Karpagam University, Coimbatore, Tamil Nadu, India
4 Meditrina Cardiac Centre, St. Thomas Hospital, Changanassery, Kerala, India
| Date of Web Publication | 14-Oct-2014 |
Correspondence Address:
Changanamkandath Rajesh
Sci Genom Labs Pvt. Ltd, Plot 43A, SDF 3rd Floor, CSEZ, Kakkanad, Kochi, Kerala
India
 Source of Support: None, Conflict of Interest: None  | 2 |
DOI: 10.4103/0971-6866.142896

 Abstract Abstract | | |
Background and Aim: This study reports the prevalence of five clinically significant variants associated with increased risk of cardiovascular disorders, and variable responses of individuals to commonly prescribed cardiovascular drugs in a South Indian population from the state of Kerala.
Materials and Methods: Genomic DNA isolated from 100 out-patient samples from Kerala were sequenced to examine the frequency of clinically relevant polymorphisms in the genes MYBPC3 (cardiomyopathy), SLCO1B1 (statin-induced myopathy), CYP2C9, VKORC1 (response to warfarin) and CYP2C19 (response to clopidogrel).
Results: Our analyses revealed the frequency of a 25 bp deletion variant of MYBPC3 associated with risk of cardiomyopathy was 7%, and the SLCO1B1 "C" allele associated with risk for statin-induced myopathy was 15% in this sample group. Among the other variants associated with dose-induced toxicity of warfarin, VKORC1 (c.1639G>A), was detected at 22%, while CYP2C9*3 and CYP2C9*2 alleles were present at a frequency of 15% and 3% respectively. Significantly, the tested sample population showed high prevalence (66%) of CYP2C19*2 variant, which determines response to clopidogrel therapy.
Conclusions: We have identified that certain variants associated with cardiovascular disease and related drug response in the five genes, especially those in VKORC1, CYP2C19 and MYBPC3, are highly prevalent in the Kerala population, with almost 2 times higher prevalence of CYP2C19*2 variant compared with other regions in the country. Since the variants chosen in this study have relevance in disease phenotype and/or drug response, and are detected at a higher frequency, this study is likely to encourage clinicians to perform genetic testing before prescribing therapy.
Keywords: Cardiovascular disease, drug response, drug toxicity, genetic testing
How to cite this article:
Mahadevan L, Yesudas A, Sajesh P K, Revu S, Kumar P, Santhosh D, Santhosh S, Sashikumar J M, Gopalakrishnan V K, Boben J, Rajesh C. Prevalence of genetic variants associated with cardiovascular disease risk and drug response in the Southern Indian population of Kerala. Indian J Hum Genet 2014;20:175-84 |
How to cite this URL:
Mahadevan L, Yesudas A, Sajesh P K, Revu S, Kumar P, Santhosh D, Santhosh S, Sashikumar J M, Gopalakrishnan V K, Boben J, Rajesh C. Prevalence of genetic variants associated with cardiovascular disease risk and drug response in the Southern Indian population of Kerala. Indian J Hum Genet [serial online] 2014 [cited 2016 Aug 23];20:175-84. Available from: http://www.ijhg.com/text.asp?2014/20/2/175/142896 |
 Introduction Introduction | |  |
Cardiovascular diseases (CVDs) have emerged as the leading cause of mortality worldwide. According to World Health Organization's 2005 report, "preventing chronic death," 30% of all global deaths (17.3 million of 58 million) are due to CVDs. [1] In India, 1.7-2.0 million deaths are attributed to CVDs annually. [2] What is especially significant is the fact that the state of Kerala has a high mortality associated with CVD (187 - >350 deaths/100,000 person/year) in the country. [3],[4]
The South Indian region, which is home to one-fifth of the total population of India, accounts for ~25% of total CVD-related deaths in the country. [2] It is estimated that by 2030, death linked to cardiovascular complications will rise to 17.9 million in India alone, which is approximately 10 times the CVD-related mortality in the United States. [5] This projected sharp increase in CVD-related death in India will necessitate implementing aggressive measures to monitor disease surveillance, as well as putting patients into effective drug therapy regimens.
The era of modern genomics has uncovered many genetic variations that predispose individuals to the risk of CVDs. [6] Further, these genome wide association studies have also predicted single nucleotide polymorphisms (SNPs) in genes that correlate with a positive response to CVD drugs. Such studies have also implicated SNPs linked to adverse drug-induced toxicity in CVD patients. Taken together, these findings provide a genetic basis to stratify patients who are likely to benefit from aggressive disease management by initiating them on drug regimens that are tailored to the patient's genetic variations. One potential caveat of this approach is that these findings cannot be readily applied to the Indian population, having been derived exclusively from the analysis of populations outside India. As an example, the prevalence of cardiomyopathy is reported to be higher in the South Asian population in comparison to the Western population. [5] Polymorphisms in genes encoding myosin binding protein C (MYBPC) and beta myosin heavy chain can lead to cardiomyopathy by compromising the structural integrity of the heart muscle and its development. Recently, a 25 bp deletion in the intron 32 of the MYBPC3 gene encoding MYBPC, a structural component of the sarcomere, has been reported to occur at an overall frequency of ~11% in the Indian population with a specific 6% prevalence in the Kerala population. [7] Until date, this deletion has been found primarily in India and South Asia, and has also been considered as a variant specific to the Indian population. [8] Polymorphisms in other genes such as SLCO1B1, CYP2C9, VKORC1 and CYP2C19 can exacerbate drug-induced cytotoxicity, or sometimes can reduce the efficacy of drugs significantly by interfering with their uptake and/or metabolism. [9],[10],[11],[12],[13],[14],[15],[16],[17],[18] In these situations, the patient needs to be put on another drug, or the dosage of the drug needs to be adjusted appropriately.
Relatively few studies have examined the presence of specific polymorphisms in genes associated with CVD risk in the Indian population. [5],[7],[8],[19] Even fewer studies have included the South Indian population, where mortality due to CVDs is reported to be higher in comparison to other regions within the country (>300 death/100,000/year). [3] Therefore, we undertook to examine the prevalence of genetic variants associated with CVD and drug response in the Southern Indian state of Kerala, by examining polymorphisms in five genes known to influence the onset of cardiomyopathy and variable responses to commonly prescribed CVD drugs, such as statin, warfarin, and clopidogrel.
 Materials and Methods Materials and Methods | |  |
Samples
Buccal mucosal cell samples were collected from 100 cardiology out-patients of Meditrina Cardiac Centre, St. Thomas Hospital, Changanassery, Kerala, India in February-March 2012. Informed consent of subjects and appropriate Ethics Committee approval was obtained after detailed explanation of the study. The 100 patients consisted of 51 males and 49 females. The average age of patients participating in the study was 66 years. Of individuals for whom clinical history data was available, 66 had been diagnosed for diabetes, 65 for hypertension and 23 were suffering from various cardiac ailments.
Isolation of genomic deoxyribonucleic acid and polymerase chain reaction amplification
Genomic DNA was isolated from the buccal swabs using the Gentra Puregene Buccal Cell Kit (QIAGEN, Germany) as per the manufacturer's protocol. DNA regions containing the MYBPC3 rs36212066 (25 bp deletion, intron 32), SLCO1B1 rs4149056 (c. 521T>C, exon 6), VKORC1 rs9923231 (c.−1639G>A, promoter), CYP2C9*13 rs72558187 (c. 269T>C, exon 2), CYP2C9*2 rs28371674 (c. 430C>T, exon 3), CYP2C9*3 rs1057910 (c. 1075A>C, exon 7), CYP2C19*3 rs4986893 (c. 636G>A, exon 4), CYP2C19*2 rs4244285 (c. 681G>A, exon 5) polymorphisms were amplified by nested polymerase chain reaction (PCR) using primers represented in [Figure 1]. A 35-cycle PCR was carried out using the following conditions: Denaturation of template DNA for the first cycle at 98°C for 30 s, denaturation at 98°C for 20 s, annealing at 64°C (MYBPC3 and SLCO1B1) or 62°C (VKORC1 and CYP2C9) or 60°C (CYP2C19) for 35 s, extension at 72°C for 1 min and extension at 72°C for 7 min. PCR was performed using a high fidelity (HF) reaction buffer containing 1.5 mM MgCl2; 50% DMSO; 10 mM of each dNTPs; 0.5 units Phusion Taq polymerase (Finnzymes, Finland); 0.5 μM of each primer and 40 ng of template genomic DNA. The outer PCR product was used as template for PCR with inner primers under the same conditions described for outer PCR. Post amplification, the PCR products were resolved on 1.5% agarose gel and eluted using GeneJET Gel extraction kit (Fermentas, Germany).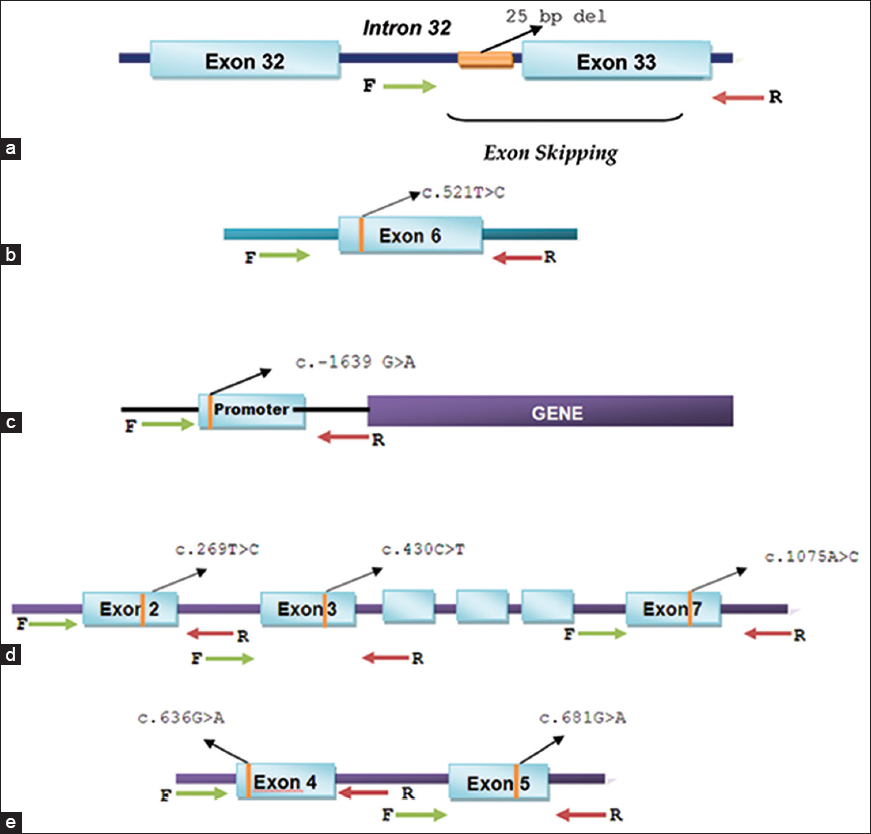 | Figure 1: Schematic representation of the strategy used to selectively amplify genetic regions harboring polymorphisms for sequencing. (a) MYBPC3 - rs36212066 (25 bp deletion, intron 32),b) SLCO1B1 - rs4149056 (c. 521T > C, exon 6),c) VKORC1 - rs9923231 (c.−1639 G > A, promoter)d) CYP2C9 alleles, - CYP2C9*13 - rs72558187 (c. 269T > C, exon 2), CYP2C9*2-rs28371674 (c. 430C > T, exon 3) and CYP2C9*3 rs1057910 (c. 1075A > C, exon 7), (e) CYP2C19 alleles, - CYP2C19*3 rs4986893 (c. 636G > A, exon 4) and CYP2C19*2 rs4244285 (c. 681G > A, exon 5)
Click here to view |
DNA sequencing and analysis for mutations
The amplified inner PCR products were sequenced on an ABI3730 × l DNA analyzer (Applied Biosystems, USA), using the ABI PRISM BigDye Terminator Cycle Sequencing Kit (Perkin-Elmer Corp., USA), following the manufacturer's instructions. The sequencing data obtained was analyzed using Sequencher, Version 5.0 (Gene Codes Corp., USA). Genotypes for the polymorphisms under study were detected using the mutation analysis software Mutation Surveyor Version 3.24 (SoftGenetics LLC, USA) where the sequences obtained were compared with the corresponding region of the gene on reference sequences downloaded from National Centre for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/).
 Results Results | |  |
MYBPC3 risk variant in Kerala population
Of the 100 samples analyzed for the presence of the MYBPC3 deletion, 7 samples were found to be heterozygous for the 25 bp deletion [Figure 2] and [Table 1]. Given that the 25 bp deletion results in a frame shifted functionally impaired protein, we failed to detect any homozygous cases in either cohorts, which are rare and has been reported previously in Indian cardiomyopathy patients (0.8%), but not in the control population. [7] The frequency of 4-7% of MYBPC3 deletion falls within the same range (2-8%) as reported from other subgroups in India. [7],[8] 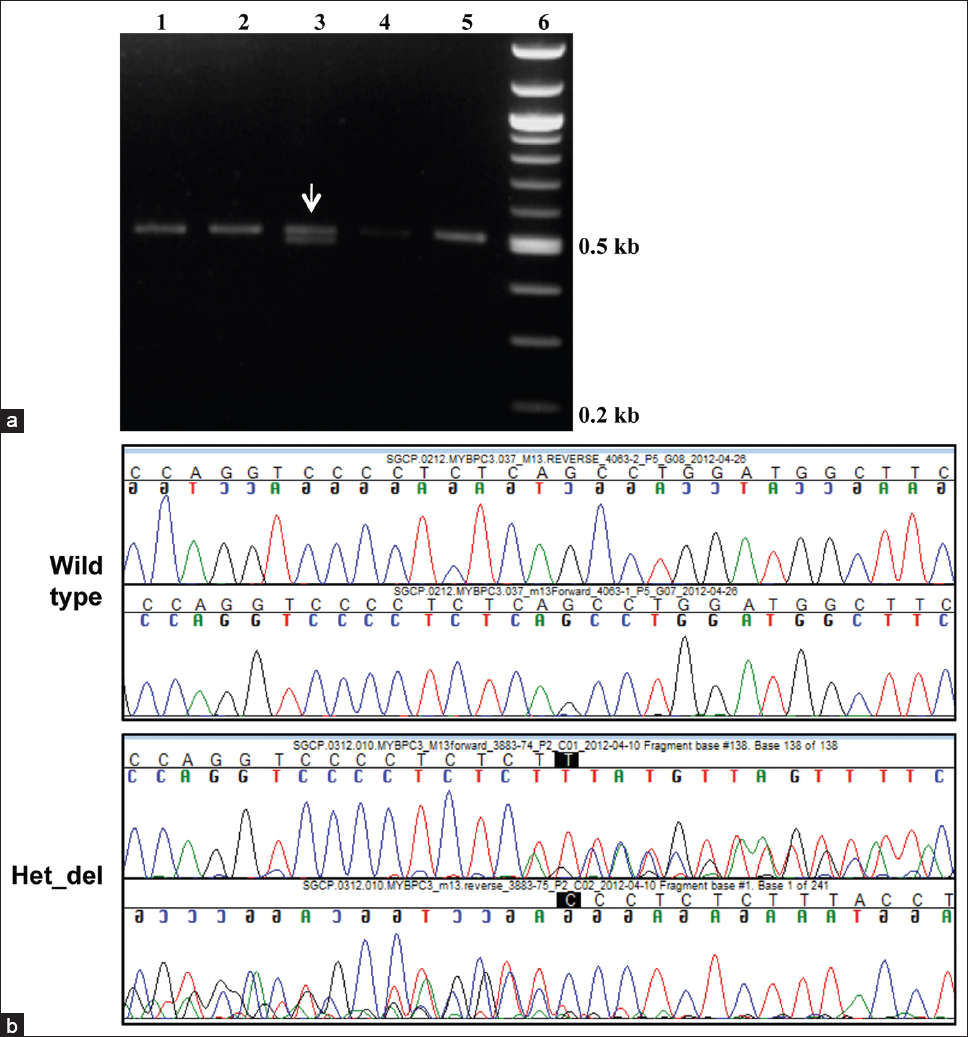 | Figure 2: Characterization of the 25 bp deletion in the MYBPC3 gene. (a) Representative agarose gel electrophoresis pattern of the PCR products obtained for the wild-type MYBPC3 (single band seen in Lanes 1, 2, 4 and 5) and a heterozygous deletion sample (two bands corresponding to the wild-type and the 25 bp deletion seen in Lane 3). The marker lane is shown on the right. (b) Representative sequence chromatogram of a heterozygous sample depicting region of heterozygous deletion (Het_del) in MYBPC3 gene viewed as mixed peaks. The control wild type chromatogram without the deletion is shown on the top
Click here to view |
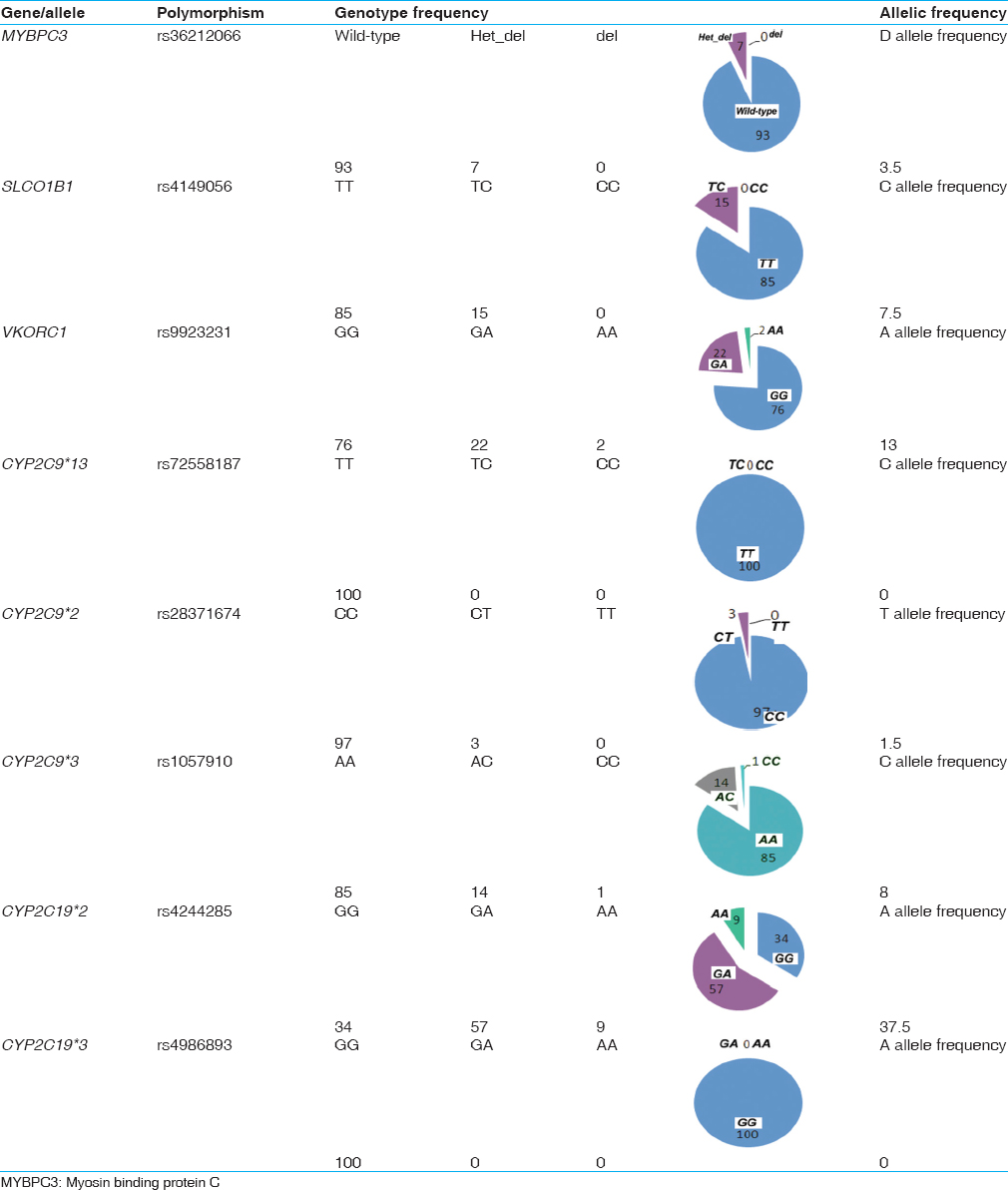 | Table 1: Genotypic and allelic frequencies for the various polymorphisms tested in the Kerala population
Click here to view |
Genetic variants exacerbating drug-induced toxicity
Several genetic polymorphisms influence drug-induced risk of cardiac malfunction. As an example, SLCO1B1, c. 521T > C; p.Val174Ala variant is associated with increased risk of statin-induced myopathy. [20],[21] Of the 100 samples analyzed, 15 samples were positive for the C allele, All which were heterozygous [Figure 3]a and [Table 1]. This distribution pattern is similar to the 15-16% reported prevalence of the C allele in populations of Caucasian decent. [10],[20],[21] A study on different ethnic groups in Brazil reported the allele to be present in highest numbers in American-Indians (28.3% heterozygous and 9.9% homozygous) and lowest in African descent subjects (5.7% and 0%) compared with Mulatto (14.9% and 2.4%) and Caucasian descent (14.8% and 4.1%). [20] 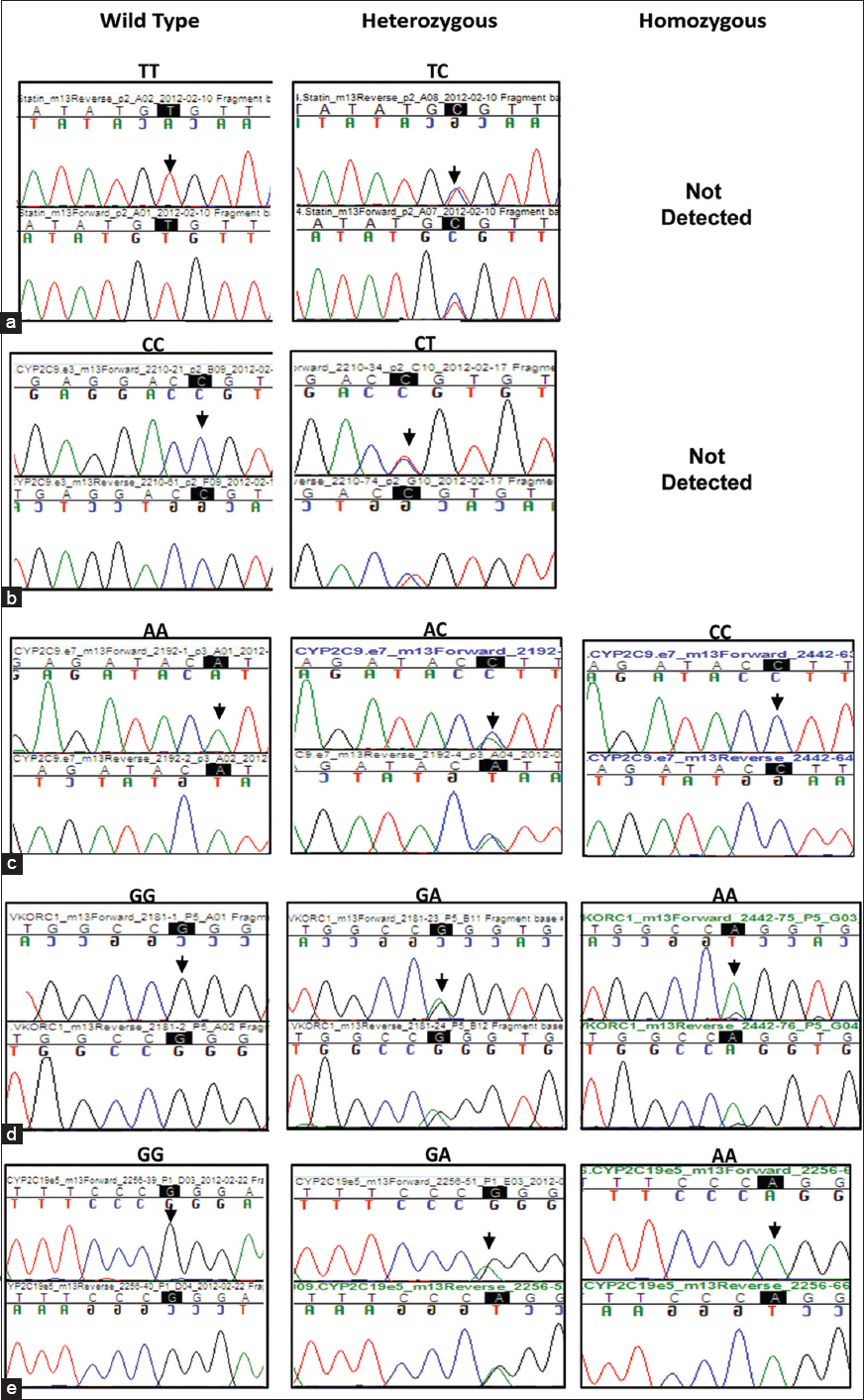 | Figure 3: Chromatogram showing the presence of different variants in the genes tested: (a) SLCO1B1 (c. 521T > C) (b) CYP2C9*2 (c. 430C > T)c) CYP2C9*3 (c. 1075A > C) (d) VKORC1 (c.-1639 G > A)e) CYP2C19*2 (c. 681G > A, exon 5). Chromatogram of the region flanking each variant is highlighted as well as indicated by an arrow over the chromatogram
Click here to view |
Interestingly, our study shows that the SLCO1B1 'C' allele was higher in females than in males, 21.5% females compared to only 7.8% males in this group of individuals [Table 2].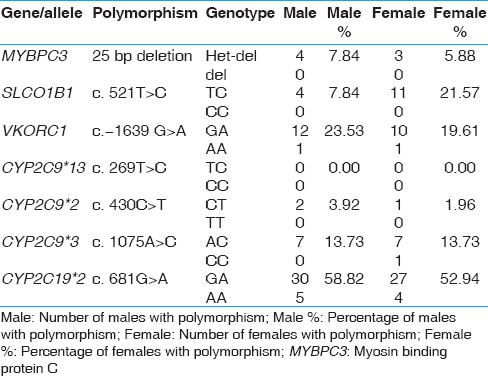 | Table 2: Gender wise distribution of polymorphisms in the Kerala study population
Click here to view |
Polymorphisms in two CYP2C9 and VKORC1 genes contribute to the cytotoxic effects of warfarin, which at high doses can cause internal hemorrhages in patients carrying certain genetic variants. Four genetic variants, three in CYP2C9 that is, CYP2C9*2, CYP2C9*3 and CYP2C9*13, (with CYP2C9*1 being the wild type allele) and one in VKORC1 are relevant to warfarin-induced cytotoxicity. Among the three CYP2C9 variants, CYP2C9*3 was detected in 15% of the study population [Figure 3]c and [Table 1]. 14% individuals were heterozygous carrying the CYP2C9*1/*3 alleles and 1% carried the homozygous (CYP2C9*3/*3) genotype. The CYP2C9*2 variant allele was heterozygous (CYP2C9*1/*2) in 3% individuals with no homozygous genotype detected [Figure 3]b and [Table 1]. The third variant CYP2C9*13 was absent in our study population. One sample was a compound heterozygote for the *2 and *3 alleles (CYP2C9*2/*3). Previous studies reported the presence of CYP2C9*3 allele in approximately 1-4% of the Asian population, while the CYP2C9*2 allele was absent. [22] The CYP2C9*2 allele is reported to be absent or very rare in East Asian populations, with the CYP2C9*1, *2 and *3 alleles reported in a Chinese Mongolian study to be 97%, 0% and 3%, respectively. [23],[24] Another study involving a broader South Indian population, showed the frequencies of CYP2C9*2 and *3 to be 4%, and 8% respectively. [25] The frequency of the CYP2C9*13 allele, not detected in the current population, was previously reported only in 1.02% of the Chinese population. [26]
Analysis of the G to A polymorphism in the 5'UTR region (-1639 position from the start) of the VKORC1 gene revealed that 22% of the individuals were heterozygous (G/A) for the variation and 2% were homozygous (A/A) [Figure 3]d and [Table 1]. This polymorphism alters transcription factor binding site upstream of the VKORC1 gene, leading to lower levels of VKORC1 protein expression, thus necessitating dosage reduction of warfarin in carriers. [16] A previous study involving 50 CVD patients from North India detected 16% prevalence in the VKORC1 (-1639G > A) allele (6 homozygous and 10 heterozygous), which is similar to what we have detected in the Kerala population. [27] By contrast, the Chinese population carry the VKORC1 variant at significantly higher frequency (82.1%, homozygous). [16]
We also detected a combination of polymorphisms in both the genes. Two patient samples (2%) carried CYP2C9*2 and VKORC1 (−1639G > A) alleles, while five samples (5%) carried CYP2C9*3 and VKORC1 (-1639G>A). These combinations were present in the heterozygous state and are expected to exacerbate drug-induced toxicity necessitating treatment with lower doses of warfarin.
We also tested two important polymorphisms in the CYP2C19 gene (CYP2C19*2 and CYP2C19*3) that results in poor conversion of clopidogrel pro-drug to its active form. [17] Genotyping assays revealed that 57% of study samples were heterozygous for the CYP2C19*2 allele (CYP2C19*1/*2) while 9% were homozygous [Figure 3]e and [Table 1]. The presence of CYP2C19*2 allele has been reported previously to be high in Asian population (29-55%) when compared with the Caucasian or African American population (13% and 18% respectively). [17],[28] The CYP2C19*3 variant was not detected in the present cohort.
 Discussion Discussion | |  |
The main objective of this study was to ascertain the frequency of genetic polymorphisms in five genes associated with the risk of cardiomyopathy, drug-induced toxicity and variable response to therapy for cardiac-related conditions in a cohort of subjects from the South Indian state of Kerala with high incidence of CVD. Based on a recent study by the Indian Genome variation consortium and other population studies on India, it has been proposed that although the country as a whole is genetically diverse, there is significant genetic homogeneity within groups related to each other by linguistic and ethnic affiliations. [29],[30] Therefore, large-scale countrywide population studies that are not properly stratified to reflect this genetic homogeneity may fail to capture correlations that exist between specific genetic variants with disease or drug response phenotype. It is very likely then, that clinically significant genetic correlations derived from one region of the country may lead to erroneous inferences of genetic association in another region. An alternative approach to overcome this uncertainty is to carry out region-specific multiple smaller studies that reveal true prevalence of region-specific genetic variants enabling clinicians to make superior inference of genetic association with the disease and help them to choose the right treatment regimens for the group as a whole. Our analyses reveal a spectrum of variations in the five clinically relevant genes at frequencies that are different from those that were detected in other regions of the country, making a strong case for requiring genetic testing within the CVD community of Kerala before considering any specific line of treatment.
The 25 bp deletion variant of MYBPC3 gene is known to influence predisposition to heart failures and in posing a lifelong threat to carriers. Earlier reports detected the presence of this deletion at a frequency of 13.8% (13% heterozygotes and 0.8% homozygotes) in pooled Indian patients with hypertrophic cardiomyopathy and only in 2.9% (heterozygous condition) of healthy control individuals. [7] Another report covering multiple countries within the Indian subcontinent showed significant variability (2-8%) between populations within this region. [8] In the Kerala population, this MYBPC3 heterozygous deletion variant was found to be present in 7% of the samples. Therefore, the higher incidence of CVD in the Kerala population cannot be attributed entirely to the presence of this deletion variant. This conclusion needs to be bolstered further with a larger cohort of cardiomyopathy patients and controls to enable identification of other risk factors associated with this disease condition in the region.
Genetic testing of polymorphisms that influence response to CVD drugs can reduce drug related toxicity and increase efficacy of treatment. Although both genetic and environmental factors contribute to drug response, it is estimated that genetics alone can account for 20-95% of the variability in drug response by impacting the absorption and metabolism of CVD drugs. [31],[32],[33] One important application of genetic testing will be to assign patients the correct dose regimen of statin, to reduce statin (Simvastatin)-induced myopathy at higher doses and prevent concurrent use of drugs like cyclosporine that inhibits statin metabolism. [9] Myopathy occurs in up to 10% of statin-treated patients and is most commonly manifested by myalgia. [34] The SLCO1B1 (c. 521T > C) C allele causes lower statin uptake in the liver and is a risk factor for statin-induced myopathy. The variant, prevalent in 15% of the Caucasian population, is responsible for more than 60% of the myopathy cases. [20],[21] We detected this variant in 15% of samples tested in the Kerala population. These individuals are likely candidates for lower doses of Simvastatin or alternate statins like pravastatin and atrovastatin or lipophilic statins such as fluvastatin as they enter the liver via passive diffusion. [33],[35] Genotyping for this polymorphism can therefore be an important clinical tool in preventing this irreversible muscle damage.
Another group of drugs whose kinetics of absorption and metabolism is affected by genetic polymorphism include oral anticoagulants such as warfarin and clopidogrel. CYP2C9 and/or VKORC1 gene variants are primary genetic determinants of warfarin dosing. Of over 24 polymorphisms known in the CYP2C9 gene, the CYP2C9*2 and CYP2C9*3 alleles are associated with an increased risk of bleeding due to inappropriate warfarin dosing. [36] Patients homozygous for the CYP2C9*3 allele (CYP2C9*3/*3) needed 3.3-fold lower mean dose of warfarin to achieve the same normalized ratio as the wild type allele (CYP2C9*1/*1), with CYP2C9*3 carriers (CYP2C9*1/*3) requiring an intermediate dose. The impact of these reduced-function alleles is related to the clearance of warfarin in vivo, which is reported to be reduced by 66% and 90% for CYP2C9*1/*3 and CYP2C9*3/*3 respectively. [23] Both the CYP2C9*2 and the CYP2C9*3 alleles are rare in the Asian population. [21],[22] A recent study detected the presence of CYP2C9*2 and *3 alleles at 4% and 8% respectively in the general South Indian population. [25] This study reports higher preponderance of the CYP2C9*3 allele, at 15% in the Kerala population while CYP2C9*2 allele was relatively rare (3%).
VKORC1 contributes to 15-30% of the variation in the dose of warfarin required. The (-1639G>A) polymorphism in the VKORC1 gene, the second primary genetic factor contributing to warfarin sensitivity accounts for over half of this variability. [14] In our cohort, 22% carry this polymorphism in a heterozygous state, which is comparable to what has been detected in another North Indian population. [27] The Chinese population on the other hand carries the polymorphism in homozygous state in 82.1% of individuals compared with 14.2% of a healthy Caucasian population indicating lower maintenance dose requirement for Chinese population. [16]
Possession of both VKORC1 and CYP2C9 variants would require a very significant (80%) dose reduction compared with the lone gene variants for prevention of adverse effects. [11],[14] Knowledge about an individual's genotype in the context of these genes therefore promises controlled use of warfarin to help avoid hemorrhagic events in cardiac patients among multiple ethnic groups studied. From the present study it becomes apparent that there is a high prevalence of three variants to adversely impact the patient treatment groups in Kerala, unless prior genotypic stratification and dose adjustments are implemented.
The third gene we analyzed contributes to the efficacy of clopidogrel therapy in patients with CVD. Previous studies have shown that CYP2C19 genetic variations account for ~30% of risk for recurrent ischemic events, with CYP2C19*2 and CYP2C19*3 loss of function variants resulting in 25-33% decreased serum levels of the active metabolite and platelet inhibition than noncarriers, doubling the risk for major adverse cardiovascular events. [17] In this study, we found that the Kerala population shows a high prevalence of CYP2C19*2 allele at 66% (57% patients heterozygous and 9% homozygous). Given the high prevalence of the CYP2C19 risk allele in the Kerala population, an assessment of this genotype will be highly beneficial in determining the dose at which clopidogrel treatment along with determination of the active compound in the blood for individuals carrying the risk alleles. In addition, alternative antiplatelet agents, like third generation thienopyridine and prasugrel should be incorporated into treatment regimens in patients with homozygous loss of function alleles.
To the best of our knowledge, this is the first study reporting the combined prevalence of clinically relevant polymorphisms in five genes in the CVD population of Kerala. Data presented in this study will enhance our understanding of the distribution of clinically important genetic variants-related to cardiac ailments and offer a preliminary rationale for the optimal choice of drugs, including arriving at a dosing regimen that will offer maximal benefit to the patients by increasing efficacy, while at the same time minimizing the risk of drug-induced toxicity. We propose that Kerala population be given routine genetic tests to detect polymorphisms in these five genes before making any decision on the course of treatment. In addition, our study also supports the use of genetic counseling to reduce the risk of heart disease in families harboring these polymorphisms.
 References References | |  |
| 1. | |
| 2. | Raghuvanshi VP, Agarwal R. Diet, lifestyle and cardiovascular disease. Int J Pharm Sci Health Care 2012;2:22-32.  |
| 3. | Soman CR, Kutty VR, Safraj S, Vijayakumar K, Rajamohanan K, Ajayan K, et al. All-cause mortality and cardiovascular mortality in Kerala state of India: Results from a 5-year follow-up of 161,942 rural community dwelling adults. Asia Pac J Public Health 2011;23:896-903.  |
| 4. | Gupta R, Joshi P, Mohan V, Reddy KS, Yusuf S. Epidemiology and causation of coronary heart disease and stroke in India. Heart 2008;94:16-26.  |
| 5. | Joshi P, Islam S, Pais P, Reddy S, Dorairaj P, Kazmi K, et al. Risk factors for early myocardial infarction in South Asians compared with individuals in other countries. JAMA 2007;297:286-94.  |
| 6. | McPherson R. From genome-wide association studies to functional genomics: New insights into cardiovascular disease. Can J Cardiol 2013;29:23-9.  [ PUBMED] |
| 7. | Dhandapany PS, Sadayappan S, Xue Y, Powell GT, Rani DS, Nallari P, et al. A common MYBPC3 (cardiac myosin binding protein C) variant associated with cardiomyopathies in South Asia. Nat Genet 2009;41:187-91.  |
| 8. | Simonson TS, Zhang Y, Huff CD, Xing J, Watkins WS, Witherspoon DJ, et al. Limited distribution of a cardiomyopathy-associated variant in India. Ann Hum Genet 2010;74:184-8.  |
| 9. | Fedacko J, Singh RB, Chaithiraphan S, Vargová V, Tomlinson B, De Meester F, et al. Clinical manifestations of adverse effects of statins, oxidative stress and possible role of antioxidants in prevention. Open Nutraceuticals J 2010;3:154-65.  |
| 10. | SEARCH Collaborative Group, Link E, Parish S, Armitage J, Bowman L, Heath S, Matsuda F, et al. SLCO1B1 variants and statin-induced myopathy - A genomewide study. N Engl J Med 2008;359:789-99.  [ PUBMED] |
| 11. | Dumas TE, Hawke RL, Lee CR. Warfarin dosing and the promise of pharmacogenomics. Curr Clin Pharmacol 2007;2:11-21.  |
| 12. | Goldstein JA. Clinical relevance of genetic polymorphisms in the human CYP2C subfamily. Br J Clin Pharmacol 2001;52:349-55.  [ PUBMED] |
| 13. | Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C. Influence of cytochrome P450 polymorphisms on drug therapies: Pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol Ther 2007;116:496-526.  |
| 14. | Limdi NA, Wadelius M, Cavallari L, Eriksson N, Crawford DC, Lee MT, et al. Warfarin pharmacogenetics: A single VKORC1 polymorphism is predictive of dose across 3 racial groups. Blood 2010;115:3827-34.  [ PUBMED] |
| 15. | Rieder MJ, Reiner AP, Gage BF, Nickerson DA, Eby CS, McLeod HL, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med 2005;352:2285-93.  |
| 16. | Yuan HY, Chen JJ, Lee MT, Wung JC, Chen YF, Charng MJ, et al. A novel functional VKORC1 promoter polymorphism is associated with inter-individual and inter-ethnic differences in warfarin sensitivity. Hum Mol Genet 2005;14:1745-51.  |
| 17. | Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, Brandt JT, et al. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med 2009;360:354-62.  |
| 18. | Serebruany VL, Steinhubl SR, Berger PB, Malinin AI, Bhatt DL, Topol EJ. Variability in platelet responsiveness to clopidogrel among 544 individuals. J Am Coll Cardiol 2005;45:246-51.  |
| 19. | Srinath Reddy K, Shah B, Varghese C, Ramadoss A. Responding to the threat of chronic diseases in India. Lancet 2005;366:1744-9.  |
| 20. | Santos PC, Soares RA, Nascimento RM, Machado-Coelho GL, Mill JG, Krieger JE, et al. SLCO1B1 rs4149056 polymorphism associated with statin-induced myopathy is differently distributed according to ethnicity in the Brazilian general population: Amerindians as a high risk ethnic group. BMC Med Genet 2011;12:136.  |
| 21. | Donnelly LA, Doney AS, Tavendale R, Lang CC, Pearson ER, Colhoun HM, et al. Common nonsynonymous substitutions in SLCO1B1 predispose to statin intolerance in routinely treated individuals with type 2 diabetes: Ago-DARTS study. Clin Pharmacol Ther 2011;89:210-6.  |
| 22. | Lee CR. CYP2C9 genotype as a predictor of drug disposition in humans. Methods Find Exp Clin Pharmacol 2004;26:463-72.  [ PUBMED] |
| 23. | Yoon YR, Shon JH, Kim MK, Lim YC, Lee HR, Park JY, et al. Frequency of cytochrome P450 2C9 mutant alleles in a Korean population. Br J Clin Pharmacol 2001;51:277-80.  |
| 24. | Nasu K, Kubota T, Ishizaki T. Genetic analysis of CYP2C9 polymorphism in a Japanese population. Pharmacogenetics 1997;7:405-9.  |
| 25. | Jose R, Chandrasekaran A, Sam SS, Gerard N, Chanolean S, Abraham BK, et al. CYP2C9 and CYP2C19 genetic polymorphisms: Frequencies in the South Indian population. Fundam Clin Pharmacol 2005;19:101-5.  |
| 26. | Yang ZF, Cui HW, Hasi T, Jia SQ, Gong ML, Su XL. Genetic polymorphisms of cytochrome P450 enzymes 2C9 and 2C19 in a healthy Mongolian population in China. Genet Mol Res 2010;9:1844-51.  |
| 27. | Rathore SS, Agarwal SK, Pande S, Mittal T, Mittal B. The impact of VKORC1-1639 G > A polymorphism on the maintenance dose of oral anticoagulants for thromboembolic prophylaxis in North India: A pilot study. Indian J Hum Genet 2011;17 Suppl 1:S54-7.  |
| 28. | Luo HR, Poland RE, Lin KM, Wan YJ. Genetic polymorphism of cytochrome P450 2C19 in Mexican Americans: A cross-ethnic comparative study. Clin Pharmacol Ther 2006;80:33-40.  |
| 29. | Indian Genome Variation Consortium. Genetic landscape of the people of India: A canvas for disease gene exploration. J Genet 2008;87:3-20.  |
| 30. | Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature 2009;461:489-94.  |
| 31. | Hitchen L. Adverse drug reactions result in 250,000 UK admissions a year. BMJ 2006;332:1109.  [ PUBMED] |
| 32. | Kamali F, Pirmohamed M. The future prospects of pharmacogenetics in oral anticoagulation therapy. Br J Clin Pharmacol 2006;61:746-51.  |
| 33. | Peters BJ, Klungel OH, de Boer A, Maitland-van der Zee AH. Genetic determinants of response to statins. Expert Rev Cardiovasc Ther 2009;7:977-83.  |
| 34. | Eckel RH. Approach to the patient who is intolerant of statin therapy. J Clin Endocrinol Metab 2010;95:2015-22.  [ PUBMED] |
| 35. | Niemi M, Pasanen MK, Neuvonen PJ. Organic anion transporting polypeptide 1B1: A genetically polymorphic transporter of major importance for hepatic drug uptake. Pharmacol Rev 2011;63:157-81.  |
| 36. | Manolopoulos VG, Ragia G, Tavridou A. Pharmacogenetics of coumarinic oral anticoagulants. Pharmacogenomics 2010;11:493-6.  |
[Figure 1], [Figure 2], [Figure 3]
[Table 1], [Table 2]
|






