|
 
 |
| ORIGINAL ARTICLE |
|
| Year : 2013 | Volume
: 1
| Issue : 1 | Page : 8-14 |
|
Prevalence of minor mutations and natural polymorphisms at the protease gene among treatment-naïve human immunodeficiency virus-1 infected individuals in Jos, Nigeria
Joseph A Anejo Okopi1, Patricia A Agaba2, Lohya Nimzing3, Placid O Ugoagwu1, Kenneth Were4, Harris Onywera4, Preston Owiti4, Newton Otecko4, Samson E Isa5, Joseph A. E. Okwori6, Solomon A Sagay7, Stephen Oguche8, John A Idoko9, Oche O Agbaji5, David E Jatau10, Steve O Olonitola10
1 AIDS Prevention Initiative in Nigeria Center, Jos University Teaching Hospital, Laboratory department, Plateau State, Nigeria
2 AIDS Prevention Initiative in Nigeria Center, Jos University Teaching Hospital, Laboratory department, Plateau State, Nigeria; Department of Family Medicine, Kenya Medical Research Institute, Kisian Kisumu, Kenya
3 AIDS Prevention Initiative in Nigeria Center, Jos University Teaching Hospital, Laboratory department, Plateau State, Nigeria; Department of Medical Microbiology, Kenya Medical Research Institute, Kisian Kisumu, Kenya
4 HIV-Research Laboratory, Kenya Medical Research Institute, Kisian Kisumu, Kenya
5 AIDS Prevention Initiative in Nigeria Center, Jos University Teaching Hospital, Laboratory department, Plateau State, Nigeria; Department of Medicine, University of Jos, Nigeria
6 Department of Microbiology, National Veterinary Research Institute, Vom
7 AIDS Prevention Initiative in Nigeria Center, Jos University Teaching Hospital, Laboratory department, Plateau State, Nigeria; Department of Obstetrics & Gynaecology, University of Jos, Nigeria
8 AIDS Prevention Initiative in Nigeria Center, Jos University Teaching Hospital, Laboratory department, Plateau State, Nigeria; Department of Pediatrics University of Jos, Nigeria
9 AIDS Prevention Initiative in Nigeria Center, Jos University Teaching Hospital, Laboratory department; Department of Medicine, University of Jos; Director General, National Agency for Control of AIDS, Abuja, Nigeria
10 Department of Microbiology, Ahmadu Bello University Zaria, Nigeria
| Date of Acceptance | 01-Jun-2013 |
| Date of Web Publication | 16-Aug-2013 |
Correspondence Address:
Joseph A Anejo Okopi
AIDS Prevention Initiative in Nigeria, Jos University Teaching Hospital, Plateau State, Nigeria
 Source of Support: This work was funded in part by the US Department of Health and Human Services, Health Resources and Services Administration (U51HA02522). The contents are solely the responsibility of the authors and do not represent the official views of the funding institutions, Conflict of Interest: None declared, Conflict of Interest: None  | Check |
 
Background: Minor mutations to protease inhibitors often occur as polymorphisms in the protease gene in non-B human immunodeficiency virus (HIV)-1 subtype among antiretroviral (ARV) treatment-naïve patients. Aims: This study sought to determine the prevalence of minor mutations occurring in the protease gene among ARV naïve HIV-1 infected patients in Jos, Nigeria. Settings and Design: We retrospectively analyzed specimen of 105 patients recruited between October 2010 and April 2011 at the HIV clinic, Jos University Teaching Hospital, Nigeria. Materials and Methods : Genotypic testing was done using an in-house genotyping system at the Kenya Medical Research Institute HIV-Research Laboratory in Kisumu, Kenya; HIV-1 viral resistance mutations assessed using Stanford HIV drug resistance database and classified using International acquired immunodeficiency syndrome (AIDS) Society (IAS)-USA list of mutations. In additional, viral subtypes were determined using REGA subtyping tool v.2.0 and CD4 levels by flow cytometry. Statistical analysis: Prevalence of mutations was computed and participants' baseline clinical and biological properties summarized by percentages for categorical variables and mean/median for quantitative variables. Results: Of the 105 samples, 100 were successfully amplified. HIV-1 subtypes identified were circulating recombinant form (CRF) - CRF02_AG (48.0%), G (41.0%), CRF06_cpx (6.0%) and A1 (5.0%). The most prevalent minor mutations among the patients occurred at positions L89M (96%), M36I (93%), K20I (77%), V82I (39%), E35Q (29%), L63P (25%) and polymorphisms at I13V (99%), R41K (86%), H69K (86%), K14R (67%). One sample presented with a major PI resistance mutation (Q58E). Conclusions: We found high rates of minor mutations and polymorphisms in circulation, possibly reflecting the drug naivety of participants. In addition, there was an evidence of transmitted drug resistance, hence targeted genetic resistance testing should be considered in national treatment guidelines. Keywords: Human immunodeficiency virus-1 drug resistance, minor mutations, protease inhibitors, treatment-naïve
How to cite this article:
Anejo Okopi JA, Agaba PA, Nimzing L, Ugoagwu PO, Were K, Onywera H, Owiti P, Otecko N, Isa SE, Okwori JA, Sagay SA, Oguche S, Idoko JA, Agbaji OO, Jatau DE, Olonitola SO. Prevalence of minor mutations and natural polymorphisms at the protease gene among treatment-naïve human immunodeficiency virus-1 infected individuals in Jos, Nigeria. J HIV Hum Reprod 2013;1:8-14 |
How to cite this URL:
Anejo Okopi JA, Agaba PA, Nimzing L, Ugoagwu PO, Were K, Onywera H, Owiti P, Otecko N, Isa SE, Okwori JA, Sagay SA, Oguche S, Idoko JA, Agbaji OO, Jatau DE, Olonitola SO. Prevalence of minor mutations and natural polymorphisms at the protease gene among treatment-naïve human immunodeficiency virus-1 infected individuals in Jos, Nigeria. J HIV Hum Reprod [serial online] 2013 [cited 2017 Apr 4];1:8-14. Available from: http://www.j-hhr.org/text.asp?2013/1/1/8/116534 |
| Introduction | |  |
The introduction of highly active antiretroviral therapy (HAART) has remarkably reduced the morbidity and mortality caused by human immunodeficiency virus (HIV)/acquired immunodeficiency syndrome (AIDS) globally. The continued success of HAART is limited by the high rates of mutation in the HIV genome. The presence of minor mutations to protease inhibitors (PIs) is common among treatment-naïve patients with HIV-1 infection, but their impact on treatment outcome is not well-understood. [1],[2] These minor mutations are also called secondary and accessory mutations. [3] The prevalence of these mutations varies greatly among treatment naïve patients and is largely dependent on subtype diversity. [4],[5] PI mutations sometimes occur naturally and others only develop in the presence of drug-pressure. Minor mutations in the protease gene do not lead to high level resistance when occurring alone, but they either improve the viral fitness or increase the drug resistance level in the presence of major mutations to PIs. The database of the genetic diversity and genotypic profile of PIs associated mutations in HIV-1 subtype B in developed countries has been well-documented. However, little is known about the prevalence of minor mutations to PIs among antiretroviral (ARV)-naïve individuals harboring non-B variants in regions with multiple circulating strains. The extremely high genetic variation of HIV-1, coupled with fast turnover of virions leads to rapid selection and the emergence of drug-resistant strains and minor mutations. [6],[7] In developing countries where access to treatment has expanded rapidly in the past decade, prevalence of resistance mutations among the untreated HIV-1 infected population is reported to be low. [8] Nigeria has the second highest burden of HIV infection in the world, second only to South Africa. The report of the 2010 HIV survey showed that the North Central Zone had the highest HIV prevalence (7.5%) of all the six geo-political zones in the country. [9] Given the increasing genetic diversity of HIV-1 and the expected increase in use of PIs, it is important to characterize protease sequences from HIV-1 subtypes occurring among drug-naive populations. In this study, the objective was to estimate the prevalence of minor mutations and polymorphisms occurring in the protease gene among HIV-1 infected treatment naïve patients in Jos, Nigeria.
| Materials and Methods | |  |
This study was carried out at the HIV clinic, Jos University Teaching Hospital (JUTH), Nigeria and some analysis at Kenya Medical Research Institute (KEMRI) HIV-Research Laboratory in Kisumu, Kenya. The entry point for all patients was either through HIV counseling and testing or referred HIV positive patients from other services within the hospital, around the community and from other neighboring states. A total of 105 HIV-1 infected treatment-naïve patients were recruited sequentially after obtaining informed consent between October 2010 and April 2011. The JUTH ethics committee approved the study protocol. A questionnaire was used to collect basic demographic data from each study participant. Criteria for inclusion in this study were patients who presented with a positive HIV test, had no previous ARV exposure and were aged 18 years and above. Blood samples were collected in ethylenediaminetetraacetic acid (EDTA)-lined containers and plasma was extracted and cryopreserved.
The CD4 + T-lymphocyte cell count was measured using Partec CyFlow Counter® (Partec GmbH, Munster Germany) using the "CD4 + Easy Count kit" according to manufacturer's instructions and as previously described. [10] Briefly, 20 μl of EDTA-anticoagulated blood was added to 20 μl of monoclonal antibodies and mixed thoroughly for 5 s and then incubated for 15 min at room temperature in the dark. Following incubation, 800 μl of no lyse dilution buffer was added to the tube and was gently mixed for 5 s. The prepared specimen was then analyzed using the Partec CyFlow counter for enumeration of CD4 + T-lymphocytes. Results were available in 2 min and recorded in cells/μl. All blood samples were processed on the same day that the blood was drawn. Plasma specimen aliquots were subsequently shipped in ice-parked containers to the KEMRI HIV-Research Laboratory within a period of 48 h for HIV-1 resistance mutation analysis and genotypic testing.
The protease region (1-99 amino acids) and part of reverse transcriptase (RT) (1-250 amino acids) regions of HIV-1 was sequenced using an in-house genotypic assay as previously described. [11] Viral ribonucleic acid (RNA) was isolated using the viral RNA mini kit (Qiagen, Hilden, Germany) and stored at −80°C until use. The primer design and modifications for this assay so as to amplify all HIV-1 group M subtypes and circulating recombinant forms (CRFs) of the pol gene region associated with resistance in the protease and RT regions of the HIV-1 genome are also described. [12] The outer primers for a one-step reverse transcription polymerase chain reaction (RT-PCR) were Prt-F1 (forward, 5'-CCTCAAATCACTCTT TGGCARCG, nucleotides (nt) 2253-2275 based on HIV-1 HXB2) and RT-R1 (reverse, 5'-ATCCCTGCATAA ATCTGACTTGC, nt 3370-3348); the reaction conditions in the ABI GeneAmp 9,700 thermocycler included 65°C for 10 min, 50°C for 45 min, 94°C for 2 min, 40 PCR cycles at 94°C for 15 s, 50°C for 20 s and 72°C for 2 min, 72°C for 10 min and left at 4°C until removal. The nested-PCR primers were Prt-F2 (forward, 5'-CTTTGGCAACGACCCCTYGTCWCA, nt 2265-2288) and RT-R2 4 μM (reverse, 5'-CTTCTGTATGTCATTGACAGTCC, nt 3326-3304). RT-PCR was performed using the RT-PRCR mixture that contained primers (Prt-M-F1 and RT-R1 each 8 μM), superscript III one-step RT-PCR system with Platinum Taq deoxyribonucleic acid (DNA) polymerase high fidelity, following the manufacturer's protocol (Invitrogen, Carlsbad, CA). Nested PCR was performed by utilizing the product of the RT-PCR, which was added to primers (Prt-F2 and RT-R2 each 8 μM), deoxynucleotide triphosphates, GeneAmp gold buffer II, 2 mM MgCl 2 , AmpliTaq Gold Low Density DNA polymerase mixture (Applied Biosystems, Foster City, CA). After initial denaturation at 94°C for 4 min, 40 cycles of PCR were performed in a GeneAmp 97,00 thermocycler with PCR conditions of 94°C for 15 s and 55°C for 20 s following an extension at 72°C for 2 min, 72°C for 10 min and 4°C until removal.
The products from nested PCR were verified by visually comparing the intensity of each sample's band to that of the DNA mass ladder's bands of known DNA quantity for expected size using electrophoresis of 1.0% agarose gel stained with 0.5 μg/ml ethidium bromide and photographed under ultraviolet transillumination. The amplified fragments were purified using the QIAquick PCR purification kit (Qiagen, Hilden, Germany) in QIAquick spin columns and were directly sequenced using BigDye Terminator v3.1 Cycle Sequencing Ready Reaction Kit with AmpliTaq DNA polymerase on an automated ABI 3,130xL Genetic Analyzer (Applied Biosystems).
The generated nucleotide sequences were manually edited using Sequencher v 5.0 (Gene Codes Corporation), which assembled the six overlapping sequence segments from the primers used to form a contiguous sequence. Sequences with frame shifts or stop codons were excluded from analysis and the quality of the generated sequences was checked using sequence quality assessment tool.
For genetic subtype determination and phylogenetic analyses; subtyping was performed using the REGA subtyping tool version 2.0 utilizing HIV-1 subtype reference sequences obtained from los Alamos database ( http://hiv-web.lanl.gov ). [13]
The PI mutations were determined using protease sequences blasted into the Stanford HIV drug resistance database and analyzed for the presence of major, minor mutations and polymorphisms at positions previously reported using the IAS-USA algorithm. [14] Data was first recorded on a predesigned paper form and subsequently transferred to Microsoft excel spread sheet. All entries were checked for possible transcription errors and the electronic data exported into the Epi Info software v3.5.3 (Centers for Disease Control and Prevention, Atlanta, Georgia) for data analysis. Baseline clinical and biological characteristics of the study subjects were summarized as percentages for categorical variables; and mean ± standard deviation or median with interquartile range (IQR) for quantitative variables. Prevalence of mutations was computed with 95% confidence interval.
| Results | |  |
Baseline characteristics
[Table 1] summarizes the baseline characteristics of these 100 patients whose specimen were successfully amplified and sequenced. Their mean age was 36.7 ± 9.5 years. Females constituted 55.0% and 69.0% were married, with the most common mode of transmission being heterosexual contact (98.0%). The median CD4 + count of patients at baseline was 186 cell/μl (IQR 12-737) and these were late presenters. A total of 45 (45.0%) had early clinical disease (World Health Organization [WHO] stages I and II) while 55.0% had advanced disease (WHO stages III and IV). A total of 46 (46.0%) patients were in a sero-discordant relationship and 14.0% of the partners of our subjects were on ARVs. Out of 105 samples in the molecular analysis, 100 were successfully amplified and sequenced. The prevalence of HIV-1 subtypes were CRF02_AG (48%), G (41%), CRF06_cpx (6%) and (A 5%). The most common subtypes were CRF02_AG and G.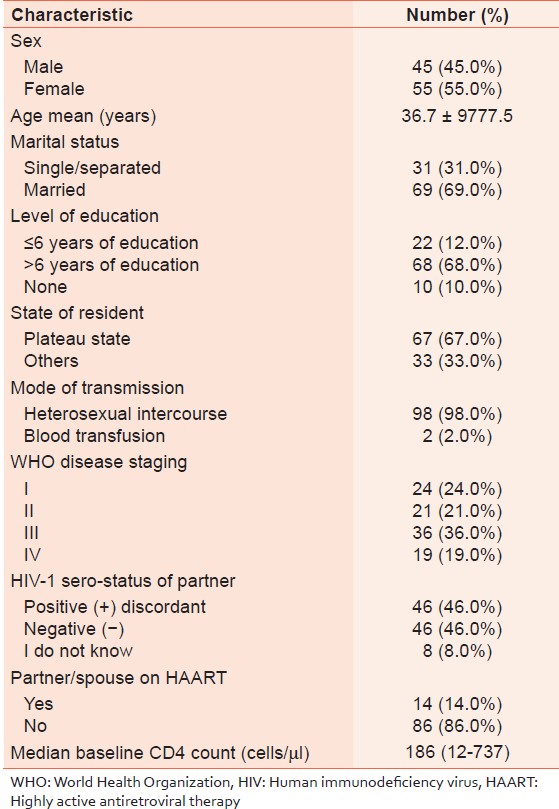 | Table 1: Socio-demographic and baseline characteristics among antiretroviral treatment-naïve patients, Jos Nigeria (n = 100)
Click here to view |
Prevalence of PI associated resistance mutations
One sample harbored a major (Q56E) PI mutation. High rates of accessory PI mutations were found at the following positions: L89M (96%), M36I (93%), K20I (77%), V82I (39%), E35Q (29%), L63P (25%), G16E (19%), I64M (18%), K20IM (15%), L10LV (13%), L10I (9%), I64L (8%), V77IV (5%), K20VI (4%), K20R (4%), K43RT (3%), I62V (3%), H69KR (2%), L89I (2%), L11LV (1%) and L33F (1%) [Figure 1]. The most common accessory mutations were L89M (96%), M36I (93%) and K20I (77%). In addition, two unusual and rare amino acid substitutions were found at positions L10M and L90V [Figure 2].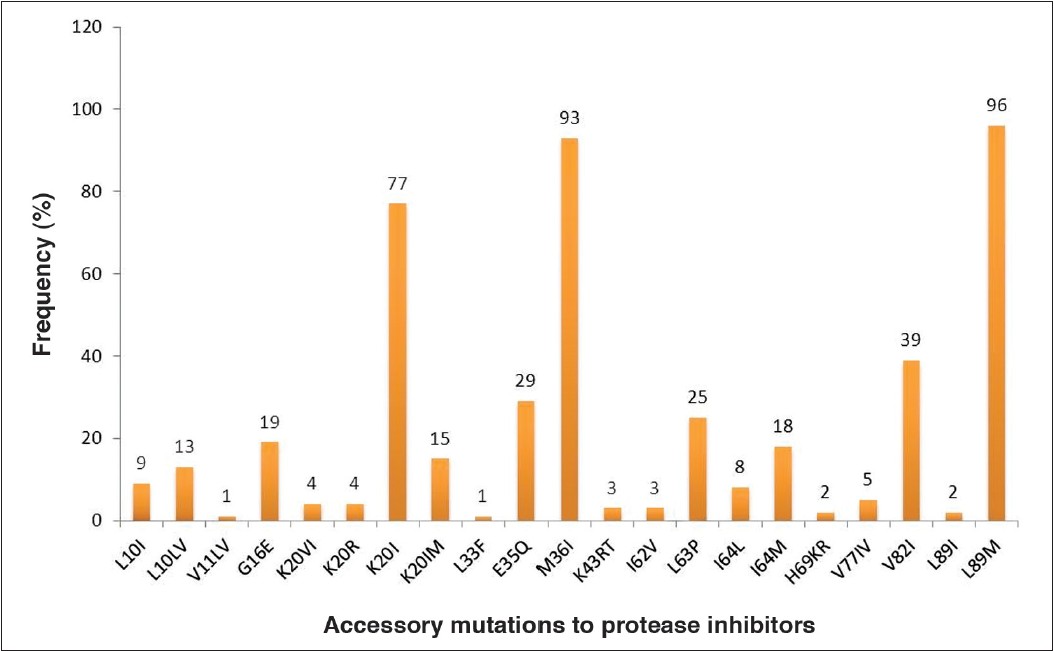 | Figure 1: Prevalence of protease accessory mutations among the antiretroviral treatment-naïve patients
Click here to view |
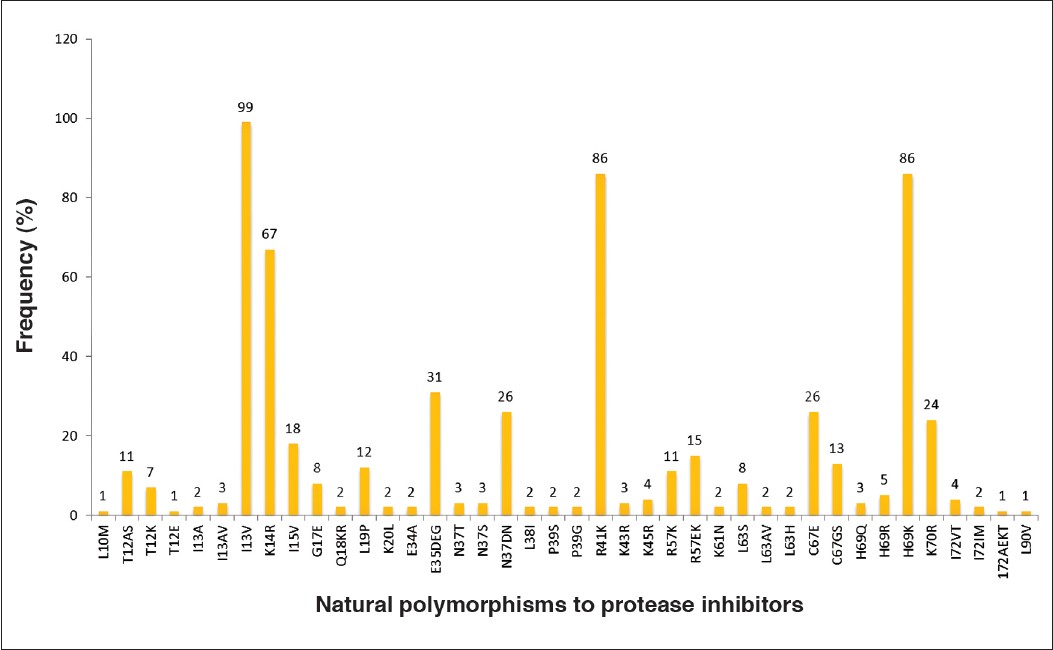 | Figure 2: Prevalence of natural polymorphisms in the protease gene among the antiretroviral treatment-naïve patients
Click here to view |
Prevalence of protease-associated polymorphisms
The prevalence of natural polymorphisms in the protease gene was high and they are represented in a deceasing order; I13V (99%), R41K (86%) H69K (86%), K14R (67%), E35DEQ (31%), N37TDN (26%), C67E (26%), K70R (24%), I15V (18%), I64IML (24%), R57EK (15%), C67GS (13%), L19P (12%), T12AS (11%), R57K (11%), L63S (8%), G17E (8%), T12K (7%) H69R (5%), I72VT (4%), K45R (4%), I13AV (3%), N37T (3%), N37S (3%), H69Q (3%), K43R (3%), E34Q (3%) and those with 2% were I13A, Q18KR, K20L, E34A, L38I, P39S, P39G, K61N, L63AV, L63H and I72IM and those with prevalence of 1% were L10M, T12E, I72AEKT, L90V (2%) and Q18KR (2%) [Figure 2].
| Discussion | |  |
The development of mutations is an important attribute of HIV biology. It is an inevitable drawback of HAART efficacy and viral response in patients under treatment and this complicates ARV regimen options during drug initiation. The result of the subtyping and phylogenetic analysis confirmed the predominance of CRF02_AG and G in Nigeria as previously reported. [15],[16],[17] The increasing prevalence of CRFO2_AG and G can be mainly associated with the phenomenon of immigrations from neighboring West-Central Africa known as the epicenter of the HIV-1 pandemic. [18],[19],[20]
The rarity of mutations to PIs is encouraging as only one sample had major drug-resistance mutation (Q58E), which is known to confer high-level resistance to tipranavir/ritonavir in drug-experienced patients. We report high rates of accessory mutations many of which are known to confer low-level resistance to atazanavir/ritonavir, Indinavir/ritonavir and lopinavir/ritonavir. [21],[22] Recent studies have shown that accessory mutations do not exhibit negative impact on therapy outcome. [23],[24] Some of the accessory mutations may not contribute significantly to decrease in drug sensitivity, but are associated with an increase in viral fitness and could influence the speed at which resistant variants are selected during therapy. [25]
High presence of polymorphisms was identified in the protease gene. The most common polymorphisms were at positions I13V (99%), R41K (86%), H69K (86%) and K14R (67%). Majority of the polymorphisms were found at very low frequencies. The effects of these mutations have not been sufficiently studied in the available variants in Nigeria, but its high presence may pose a challenge in future. It is not known whether non-B viruses with high prevalence of some amino acid substitutions at positions 13, 20, 36 and 89 may have the capacity to evolve viral fitness as compared with subtype B viruses. The need for long-term evaluation of drug efficacies in closely monitored human cohorts would determine fully the clinical significance of these substitutions. This known baseline substitutions patterns of protease amino acid in the prevailing non-B subtype variants may be useful in formulating policy and encouraging epidemiological studies. As evidenced in this study, there exist naturally occurring mutations that may vary in different epidemics, hence for public health importance, drug therapy and options may need to be adjusted to local viral resistance profiles for optimum treatment benefits. In conclusion, the study provides information on circulating HIV-1 genetic variability and resistance mutations associated with PIs among ARV treatment naïve patients at baseline. Although the drug-naïve of these study participants was limited to self-report, which may not have been free of reporter-bias, these results may indicate that routine drug-resistance testing may be currently unnecessary for every patient considered for placement on PI-based ARV therapy. However, from public health perspective and due to the increasing number of HIV-1 infected individuals with decreasing access to treatment and availability of ARVs for infected population, these data may have important implications in predicting the evolutionary trends of resistant variants under drug selection. Larger studies and continuous surveillance among drug-naïve populations are required, in order to recognize early significant changes, which may occur in the future.
| Acknowledgments | |  |
We are deeply indebted to patients who agreed to participate in this study. We thank the Director and staff of HIV-1 drug resistance testing laboratory, Kenya Medical Research Institute Kisian Kisumu, Dr. Clement Zeh who permitted the training on genotypic resistance test and analyses of the samples. We are also grateful for the support of the following people: Prof. Innocent Ujah mni, Director General Nigeria Institute of Medical Research Lagos, Mr. Joshua Adetunji of Medicom Laboratories Jos and Dr. Prosper Okonkwo Chief Executive Officer, AIDS prevention initiative in Nigeria (APIN). Ramyil Seljul, Funmilayo Moulton and Titus Obadiah were instrumental in overseeing sample collection, transport and processing. We are also grateful to APIN, Jos University Teaching Hospital, Jos for permission to use the data.
| References | |  |
| 1. | Kinomoto M, Appiah-Opong R, Brandful JA, Yokoyama M, Nii-Trebi N, Ugly-Kwame E, et al. HIV-1 proteases from drug-naive West African patients are differentially less susceptible to protease inhibitors. Clin Infect Dis 2005;41:243-51. 
[PUBMED] |
| 2. | Velazquez-Campoy A, Todd MJ, Vega S, Freire E. Catalytic efficiency and vitality of HIV-1 proteases from African viral subtypes. Proc Natl Acad Sci U S A 2001;98:6062-7. 
[PUBMED] |
| 3. | Shafer RW, Schapiro JM. HIV-1 drug resistance mutations: An updated framework for the second decade of HAART. AIDS Rev 2008;10:67-84. 
[PUBMED] |
| 4. | Rhee SY, Kantor R, Katzenstein DA, Camacho R, Morris L, Sirivichayakul S, et al. HIV-1 pol mutation frequency by subtype and treatment experience: Extension of the HIVseq program to seven non-B subtypes. AIDS 2006;20:643-51. 
[PUBMED] |
| 5. | Vergne L, Stuyver L, Van Houtte M, Butel C, Delaporte E, Peeters M. Natural polymorphism in protease and reverse transcriptase genes and in vitro antiretroviral drug susceptibilities of non-B HIV-1 strains from treatment-naive patients. J Clin Virol 2006;36:43-9. 
[PUBMED] |
| 6. | Coffin JM. HIV population dynamics in vivo: Implications for genetic variation, pathogenesis, and therapy. Science 1995;267:483-9. 
[PUBMED] |
| 7. | Condra JH, Schleif WA, Blahy OM, Gabryelski LJ, Graham DJ, Quintero JC, et al. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 1995;374:569-71. 
[PUBMED] |
| 8. | Hamers RL, Wallis CL, Kityo C, Siwale M, Mandaliya K, Conradie F, et al. HIV-1 drug resistance in antiretroviral-naive individuals in sub-Saharan Africa after rollout of antiretroviral therapy: A multicentre observational study. Lancet Infect Dis 2011;11:750-9. 
[PUBMED] |
| 9. | National Agency for the Control of AIDS FRoN. Global AIDS Response Progress Report (GARPR). InAbuja, Nigeria, 2012. Available from: http://www.unaids.org/en.or/dataanalysis/knowyourresponse/countryprogressreports/2012countries/NigerNi%202012%20GARPR%20Report%20Revised.pdf. [Last accessed on 2012 Jan 15]. 
|
| 10. | Zijenah LS, Kadzirange G, Madzime S, Borok M, Mudiwa C, Tobaiwa O, et al. Affordable flow cytometry for enumeration of absolute CD4 + T-lymphocytes to identify subtype C HIV-1 infected adults requiring antiretroviral therapy (ART) and monitoring response to ART in a resource-limited setting. J Transl Med 2006;4:33. 
[PUBMED] |
| 11. | Yang C, McNulty A, Diallo K, Zhang J, Titanji B, Kassim S, et al. Development and application of a broadly sensitive dried-blood-spot-based genotyping assay for global surveillance of HIV-1 drug resistance. J Clin Microbiol 2010;48:3158-64. 
[PUBMED] |
| 12. | McNulty A, Jennings C, Bennett D, Fitzgibbon J, Bremer JW, Ussery M, et al. Evaluation of dried blood spots for human immunodeficiency virus type 1 drug resistance testing. J Clin Microbiol 2007;45:517-21. 
[PUBMED] |
| 13. | de Oliveira T, Deforche K, Cassol S, Salminen M, Paraskevis D, Seebregts C, et al. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics 2005;21:3797-800. 
[PUBMED] |
| 14. | Johnson VA, Calvez V, Gunthard HF, Paredes R, Pillay D, Shafer RW, et al. Update of the drug resistance mutations in HIV-1: March 2013. Top Antivir Med 2013;21:6-14. 
[PUBMED] |
| 15. | Chaplin B, Eisen G, Idoko J, Onwujekwe D, Idigbe E, Adewole I, et al. Impact of HIV type 1 subtype on drug resistance mutations in Nigerian patients failing first-line therapy. AIDS Res Hum Retroviruses 2011;27:71-80. 
[PUBMED] |
| 16. | Lar P, Lar N, Bemis K, Jelpe J, Enzyguirre L, Ayuba L, et al. Subtype-specific patterns in HIV type 1 reverse transcriptase and protease in Oyo State, Nigeria: Implications for drug resistance and host response. Afr J Biotechnol 2007;6:1892-7. 
|
| 17. | Ojesina AI, Sankalé JL, Odaibo G, Langevin S, Meloni ST, Sarr AD, et al. Subtype-specific patterns in HIV Type 1 reverse transcriptase and protease in Oyo State, Nigeria: Implications for drug resistance and host response. AIDS Res Hum Retroviruses 2006;22:770-9. 
|
| 18. | Imamichi H, Koita O, Dabitao D, Dao S, Ibrah M, Sogoba D, et al. Identification and characterization of CRF02_AG, CRF06_cpx, and CRF09_cpx recombinant subtypes in Mali, West Africa. AIDS Res Hum Retroviruses 2009;25:45-5. 
[PUBMED] |
| 19. | Njai HF, Gali Y, Vanham G, Clybergh C, Jennes W, Vidal N, et al. The predominance of human immunodeficiency virus type 1 (HIV-1) circulating recombinant form 02 (CRF02_AG) in West Central Africa may be related to its replicative fitness. Retrovirology 2006;3:40. 
[PUBMED] |
| 20. | Sankale JL, Hamel DJ, Thakore S, Gueye-Ndiaye A, Eisen G, Olaleye DO, et al. Origin and diversification of CRF02_AG in West Africa. The XV International AIDS Conference: Abstract no. ThOrA1361; 2004. 
|
| 21. | De Luca A, Cozzi-Lepri A, Perno CF, Balotta C, Di Giambenedetto S, Poggio A, et al. Variability in the interpretation of transmitted genotypic HIV-1 drug resistance and prediction of virological outcomes of the initial HAART by distinct systems. Antivir Ther 2004;9:743-52. 
[PUBMED] |
| 22. | Holguín A, Soriano V. Resistance to antiretroviral agents in individuals with HIV-1 non-B subtypes. HIV Clin Trials 2002;3:403-11. 
|
| 23. | Champenois K, Deuffic-Burban S, Cotte L, André P, Choisy P, Ajana F, et al. Natural polymorphisms in HIV-1 protease: Impact on effectiveness of a first-line lopinavir-containing antiretroviral therapy regimen. J Med Virol 2008;80:1871-9. 
|
| 24. | Scherrer AU, Ledergerber B, von Wyl V, Böni J, Yerly S, Klimkait T, et al. Minor protease inhibitor mutations at baseline do not increase the risk for a virological failure in HIV-1 subtype B infected patients. PLoS One 2012;7:e37983. 
|
| 25. | Theys K, Deforche K, Vercauteren J, Libin P, van de Vijver DA, Albert J, et al. Treatment-associated polymorphisms in protease are significantly associated with higher viral load and lower CD4 count in newly diagnosed drug-naive HIV-1 infected patients. Retrovirology 2012;9:81. 
[PUBMED] |
[Figure 1], [Figure 2]
[Table 1]
|