Event
October 11-14, 2010 Odaiba, Tokyo, Japan
Biocuration 2010
The Conference of the International Society for Biocuration
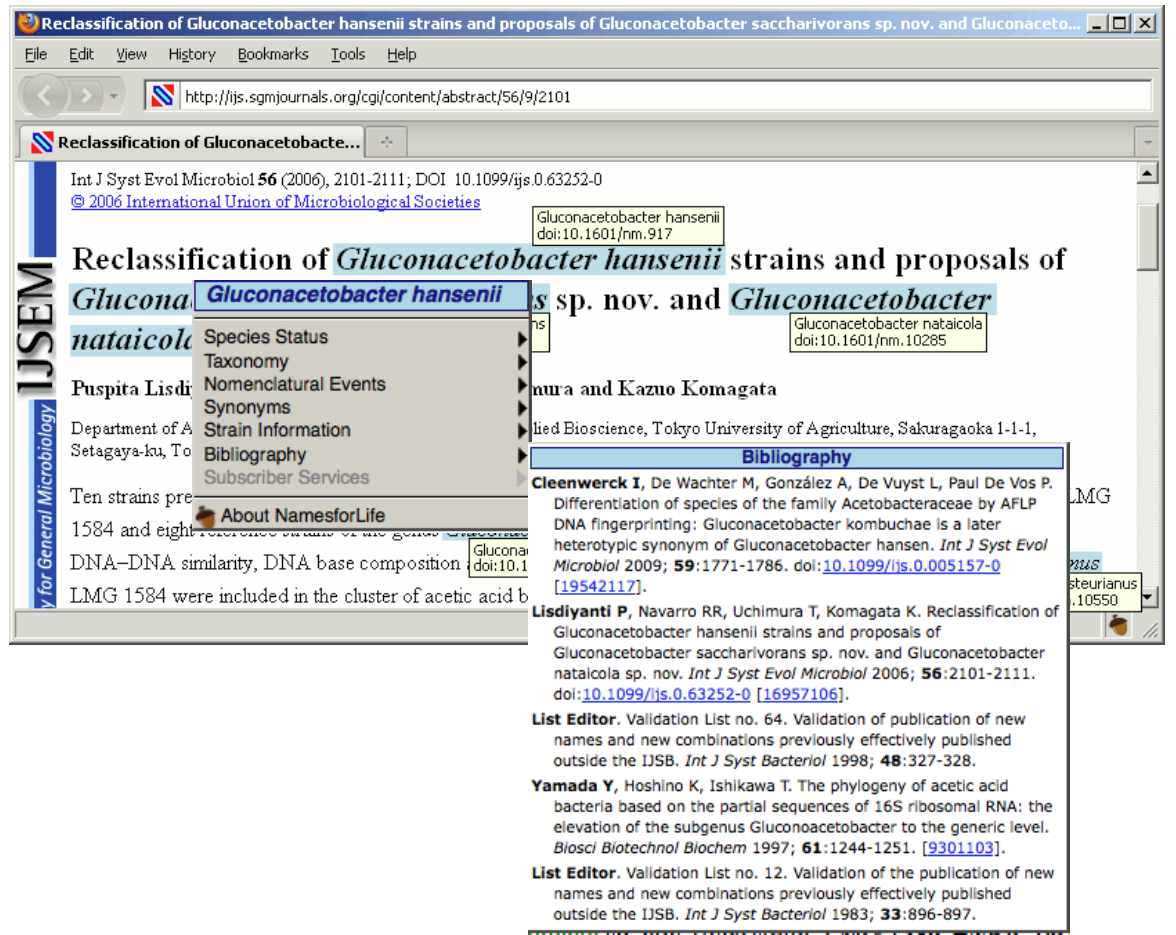
To assist those confronted with ambiguous names (which not only includes researchers but clinicians, manufacturers, patent attorneys, and others who use biological data in their routine work), we developed a generalizable semantic model that represents names, concepts, and exemplars (representations of biological entities) as distinct objects. By identifying each object with a Digital Object Identifier (DOI), it becomes possible to place forward-pointing links in the published literature, in databases, and vector graphics that can be used as part of a mechanism for resolving ambiguities, thereby “future proofing” a nomenclature or terminology. A full implementation of the N4L model for the Bacteria and Archaea was released in April, 2010. The system is professionally curated and represents a Tier III resource in Parkhill’s view of bioinformatic services (Genomic information infrastructure after the deluge, Parkhill et al. 2010). A variety of tools and web services have been developed for readers, publishers, and others (N4L Guide, N4L Autotagger, N4L Semantic Search, N4L Taxonomic Abstracts) and we are incorporating other taxonomies into the N4L data model, as well as adding additional phenotypic, genotypic, and genomic information to the existing exemplars to add greater value to end users.
[permalink] Posted October 1, 2010.