Serotonin 5-HT2 Receptors: Molecular and Genomic Diversity
Abstract
Genetic and molecular events regulate the creation of variants of 5-HT2A and 5-HT2C receptors whose diversity has important functional significance. Overall sequence identity between the 5-HT2A and 5-HT2C receptors is quite high and it is not surprising that the mechanisms of regulation for these two receptors are similar. There is, however, one striking difference: RNA editing, a mechanism for generating molecular diversity by altering the genetic code at the level of RNA. The 5-HT2C receptor is the only G protein–coupled receptor known to be edited. We examine the process of editing, as well as more common genetic modifications, at the cellular level and in human disease.
Introduction
Serotonin (5-hydroxytryptamine, 5-HT) is a phylogenetically ancient molecule. In mammals, its principal role is to regulate neural transmission. Serotonin neurons are localized in brainstem raphe nuclei and have extensive projections throughout the forebrain and into the spinal cord. The major source of 5-HT in the body is the intestine, where it acts locally and is also released into the bloodstream. Given its broad distribution, it is not surprising that 5-HT influences many spheres of mammalian physiology, from the cardiovascular and gastrointestinal systems to centrally mediated activities, such as appetite, aggression, sexual behavior, mood, cognition, learning, and memory. These diverse actions of 5-HT are mediated by multiple receptors––fourteen have been cloned and are classified into seven families (1) . The complex biology of serotonin provides fertile ground for drug therapy and new drug development. Indeed, 5-HT has been implicated in an amazing variety of human diseases ranging from major psychiatric diseases, such as schizophrenia and major depressive illness, to personality disorders such as addictive behaviors, anxiety, and eating disorders.
This review focuses on two receptors: the 5-HT2A and 5-HT2C subtypes. These closely related receptors are members of the rhodopsin family of G protein–coupled receptors (GPCRs), and both receptors couple to multiple intracellular signaling pathways (2) . The classical signal transduction pathway is Gq -coupled activation of phospholipase C (PLC) although both also activate phospholipase D and phospholipase A2 by interacting with additional G proteins. Both the 5-HT2A and 5-HT2C receptors are modified posttranscriptionally by RNA splicing, a common mechanism for achieving protein diversity. RNA editing, on the other hand, is a less common process for generating molecular diversity. In fact, the 5-HT2C receptor is the only GPCR known to be edited. RNA editing of the 5-HT2C receptor generates functionally distinct protein variants by altering the genetic code at the level of RNA. 5-HT2A and 5-HT2C receptors achieve genetic diversity through single nucleotide polymorphisms. This review examines human genetic studies of 5-HT2A and 5-HT2C receptors in the context of the relationship between the biological effects of the genetic and molecular variations and the biological basis of the disease.
Molecular Diversity: Regulation During RNA Processing
Mechanisms for generating molecular diversity exist at multiple levels of protein processing, including RNA splicing and RNA editing. Recent evidence suggests that normal RNA editing is modified in brain diseases and by drugs, making the study of this molecular process especially interesting.
RNA Splicing
Genes for both the 5-HT2A receptor and the 5-HT2C receptor contain introns, which must be removed by RNA splicing prior to RNA translation. Because common or alternative RNA splice sites can be utilized in the removal of introns from nascent RNA, multiple protein isoforms may arise from a single genetic locus. So far, all of the products of alternative RNA splicing of 5-HT2A and 5-HT2C receptors encode pharmacologically inactive proteins. For example, the 5-HT2A receptor undergoes alternative splicing that produces a variant RNA with a 112-base insert, resulting in a frame shift and premature stop codon (3) . This transcript encodes a nonfunctional, truncated protein that terminates in the fourth transmembrane region. Although it is possible that the truncated protein might act in a dominant-negative manner to regulate the function of the full-length protein, careful observations indicate that this does not occur (3) .
One alternatively spliced variant of the 5-HT2C receptor, with a 96-bp deletion occurring at the junction of exon II and III, results in a protein that is truncated near the putative junction of the second intracellular loop and the fourth transmembrane domain (4) . The truncated mRNA is expressed in parallel with the full-length mRNA throughout the brain. Studies of the function of the truncated protein (expressed alone or in combination with the full-length receptor) suggest that the splice variant is inactive. Wang et al. (5) identified a second splice variant that also encodes a truncated protein with no apparent function.
RNA Editing: Historical Overview
RNA editing was initially described as a phenomenon in which uridine residues were inserted and deleted from mitochondrial RNAs of kinetoplastid protozoa (6) . Currently, this process is generally defined as any event that changes the coding potential of primary RNA transcripts by mechanisms other than splicing. The most common form of editing, involving substitution of nucleotides, is broadly distributed in nature, from viruses to mammals (7) . The nucleotide substitution consists of either cytidine-to-uridine (C-to-U) or adenosine-to-inosine (A-to-I) alterations, which may result in altered coding potential of the mRNA and altered function of the encoded protein.
The first example of RNA editing on a nucleus-encoded mRNA was found in apolipoprotein B (ApoB ) transcripts from mouse intestine. Cytidine at the C4 position of ApoB pre-mRNA is converted to uridine by cytidine deaminase, changing a glutamine codon (CAA) into a stop codon (UAA), thereby producing a novel ApoB protein isoform with distinct physiological properties (8) . A tripartite regulatory sequence (i.e., efficiency element, spacer, and mooring sequences) surrounding the edited cytidine residue along with a multiprotein complex called an editosome is required for this modification to take place (9) . Although C-to-U changes are easily detected, A-to-I editing is more difficult to identify and involves enzymatic deamination of adenosine residues at the C6 position of the purine ring to inosines (10) . Because inosines have base-pairing properties similar to that of guanosines, inosines are recognized and translated as guanosine by the decoding ribosomes (11, 12) . Additionally, A-to-I editing requires short complementary RNA sequences embedded in an adjacent intron to form an imperfect RNA duplex with the exonic target sequence that contains the adenosine to be deaminated (13) . Given that A-to-I editing occurs in introns as well as in exons, and that intron–exon base-pairing interactions are required, it is clear that RNA editing is a nuclear event that precedes RNA splicing (11) .
The α -amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) subtype of the glutamate receptor is composed of four subunits, GluR-A, -B, -C, and -D, which form homomeric or heteromeric ligand-gated ion channels (14) . AMPA receptors containing the GluR-B subunit are impermeable to Ca2+ , whereas those without GluR-B are permeable (15) . Comparisons of genomic and cDNA sequences of transcripts encoding the GluR-B subunit reveal adenosine-to-guanosine changes. In fact, it is the RNA editing of GluR-B that controls its calcium permeability; editing of a specific arginine to glutamine within the second transmembrane (TM2) region of the protein renders the channel impermeable to calcium. Nearly all GluR-B transcripts are edited, thus explaining the calcium impermeability of this ion channel (16) . The functions of heteromeric kainate receptors also appear to be regulated by RNA editing of specific subunits (i.e., GluR-5 and GluR-6) (16) .
RNA Editing of the 5-HT2CReceptor
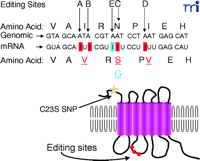
Editing of the 5-HT2C receptor was discovered by comparing sequences from genomic DNA and cDNAs from the rat striatum; four A-to-G discrepancies were identified in rat brain (17) . The cDNA library predicted the presence of valine (GTG), serine (AGT), and valine (GTT) (5-HT2C-VSV ) at positions 157, 159, and 161, respectively, whereas genomic DNA predicted isoleucine (ATA), asparagine (AAT), and isoleucine (ATT) (5-HT2C-INI ) at these three positions. As a result, it was proposed that the four pertinent adenosine residues in the 5-HT2C receptor pre-mRNA (sites termed A, B, C, and D) (Figure 1⇓) were converted into inosines in the mature mRNA by a process analogous to the editing of AMPA receptor subunits. Further analysis of cDNA sequences isolated from rat brain revealed the tissue-specific expression of seven major 5-HT2C receptor isoforms that are encoded by eleven distinct RNA species. The most prominent variant found in whole rat brain extracts is encoded primarily by editing at the A, B, and D sites and expresses the 5-HT2C-VNV receptor protein. On the other hand, editing at the A and B sites was less prominent in choroid plexus, thus generating higher levels of the 5-HT2C-INI , 5-HT2C-INV , and 5-HT2C-ISV receptor variants. In the human brain, a novel editing site was found and termed E (previously named C’) (Figure1⇓). This modification occurs at codon 158, converting an asparagine into a glycine, and results in the fully edited 5-HT2C-VGV protein (18) . These editing events take place in the putative second intracellular loop of the receptor (17) , thereby altering the coding potential in a region implicated in receptor–G protein coupling (18–20) .
5-HT2C R editing sites. Nucleotide sequence and predicted amino acid sequence alignments between the 5-HT2CR genomic DNA and mRNA sequences. A-I changes are shown in boxes (red), site of editing unique to human 5-HT2CR (blue box). The top row of amino acids indicates the predicted unedited residues, whereas the bottom row shows changes resulting from editing (underlined). The schematic diagram showcases the positions of the Cys23Ser SNP (yellow star) and editing sites (red circles) in the 5-HT2CR.
RNA Editing of the 5-HT2C Receptor: Molecular Mechanisms
The family of adenosine deaminases that act on RNA (ADARs) consists of three members: ADAR 1 and 2, which are ubiquitously expressed, and ADAR3, which is highly enriched in brain. These enzymes bind specifically to double-stranded RNA and catalyze the conversion of adenosine to inosine by hydrolytic deamination. Analysis of the rat 5-HT2C receptor gene, in the region surrounding the potential editing sites, suggests the existence of an imperfect inverted repeat forming a putative RNA duplex between the 3′ end of exon 3 and the proximal region of intron 3 (21) . In order to study the molecular mechanisms of the post-transcriptional modification of 5-HT2C receptor transcripts, Burns et al. (17) developed an in vitro editing system using rat brain nuclear extracts fractionated by cation-exchange chromatography, and a 5-HT2C receptor RNA substrate labeled with [α -32 P] adenosine 5’-triphosphate. These experiments revealed two peaks of inosine that eluted with two distinct peaks of editing activity. Activity of the first peak was responsible for editing 5-HT2C receptor transcripts at sites A, B, and C and coeluted with the activity of ADAR1, which had been shown previously to modify the B subunit of AMPA glutamate receptors (22) . The second peak of activity specifically modified the D site of the transcripts and coeluted with the activity of ADAR2, the enzyme responsible for modifying the Q/R site of GluR-B (23) .
RNA Editing of the 5-HT2C Receptor: Functional Consequences
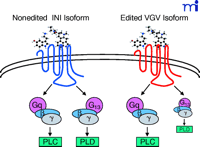
Because the editing of the 5-HT2C receptor occurs in a region involved in G protein coupling, it was hypothesized that this posttranscriptional modification may alter receptor function. In order to test the functional consequences of RNA editing, NIH-3T3 fibroblasts were transfected with cDNAs encoding different 5-HT2C receptor isoforms, and the resulting ability to activate phospholipase C (PLC) was assayed. The rat 5-HT2C-VSV isoform displayed 10–15-fold lower potency upon 5-HT stimulation compared to the unedited 5-HT2C-INI isoform. Affinity changes, desensitization, and spare receptors were all ruled out as possible explanations for these differences, leaving reduced receptor–G protein coupling as the most likely mechanism for the alteration in potency (17) . The pattern of editing in human brain differs from that found in rat, principally due to increased editing at the C and E positions (18, 24) . The most prevalent isoform in human brain is 5-HT2C-VSV, whereas 5-HT2C-VNV is most abundant in rat. The human 5-HT2C-VSV receptor exhibits a 5-fold increase in EC50 for 5-HT when compared to the 5-HT2C-INI receptor, whereas the human 5-HT2C-VGV variant shows an even greater increase in EC50 (29-fold). The 5-HT2C-INI receptor exists in a high- and low-affinity state for 5-HT, whereas the 5-HT2C-VGV receptor has lost the agonist high-affinity state (18) . The 5-HT2C-VGV receptor is also unable to stimulate inositol phosphate formation in the absence of agonist, a property referred to as constitutive activity that has been well established to occur at the rat and human 5-HT2C -INI receptors (25) . Price and Sanders-Bush (26) later showed that editing also delays agonist-stimulated calcium release. Taken together, these functional studies in cell lines show that unedited 5-HT2C receptors couple more efficiently to Gq protein even when no agonist is present, suggesting that editing may be a way to decrease tone and signaling in certain brain regions. More recently, studies of the profile of G protein coupling utilizing a high-throughput functional assay have demonstrated not only that Gq coupling efficiency is reduced, but also that the pattern of G-protein coupling is altered (27). The 5-HT2C-INI receptor isoform couples to G13 as well as Gq/11 ; however, the 5-HT2C-VGV and 5-HT2C-VSV receptor isoforms fail to couple to G13 , which results in altered intracellular signaling (Figure 2⇓).
Functional consequences of RNA editing. RNA editing alters the G protein–coupling profile of the 5-HT2C receptor with subsequent fine-tuning of downstream signals. INI (Ile, Asn, Ile) and VGV (Val, Gly, Val) refer to the amino acid residues at position 156, 158, and 160 (see text for additional details). PLC, phospholipase C; PLD, phospolipase D.
As an initial strategy for evaluating the clinical significance of RNA editing, cell lines expressing variant 5-HT2C receptor isoforms were tested with the hallucinogenic drug lysergic acid diethylamide (LSD) or antipsychotic drugs. A marked reduction in the efficacy of LSD and antipsychotics was found at 5-HT2C-VGV receptors, suggesting a possible role for editing in the etiology and treatment of schizophrenia (18, 28) . Initial studies of antipsychotic drugs suggested that a reduction in the constitutive activity of the non-edited INI isoform differentiated atypical from typical antipsychotics (29) ; however, a more recent study failed to support this hypothesis (30) . No differences in editing efficiencies were found when comparing schizophrenic patients versus controls, although there was significant elevation of editing at the A-site in a subset of psychiatric patients that committed suicide (28) . A more recent study showed that depressed suicide victims have increased editing at the E-site in the prefrontal cortex (31) . In contrast, Sodhi et al. (32) reported that editing of the 5-HT2C receptor was reduced in a small sample of schizophrenia patients. Clearly, there is a need for additional studies in larger patient samples and specific brain areas. Recent evidence suggests that 5-HT2C receptor editing is modulated by changes in 5-HT neurotransmission, including 5-HT depletion and treatment with agonists and antagonists (33) . These findings lend further support to the hypothesis that RNA editing is an important process that regulates synaptic input by fine tuning optimal signaling. This in turn may lead to novel drug treatments for psychotic patients.
Genomic Diversity: Regulation at the Level of Gene Structure
Common genetic variation might significantly contribute to increased risk for common diseases––termed the common disease-common variant hypothesis (34) . If this hypothesis is accurate, then normal genetic variation within serotonin receptors could increase the likelihood of developing a psychiatric disorder linked to serotoninergic pathways, such as depression or schizophrenia. This section reviews the clinical genetic association studies of the 5-HT2A/2C receptors as well as studies of the impact of these genetic alterations on the structure and function of the receptors.
Single Nucleotide Polymorphisms
Single nucleotide polymorphisms (SNPs), substitutions of the most common nucleotide for a differing nucleotide within genomic DNA, are the most common type of genetic variation and occur at a frequency of approximately 1% in the general population (34) . Within the coding region of a gene, SNPs that cause a change in the encoded amino acid (non-synonymous SNP) may have deleterious consequences for protein folding, producing an unstable conformation of the protein that is retained within the endoplasmic reticulum (35) . Non-synonymous SNPs may also interfere with plasma membrane retention mechanisms, such as palmitoylation and glycosylation. Finally, amino acid substitutions may be detrimental to the proper functioning of a protein. For example, non-synonymous SNPs in GPCRs could modify the binding pocket of the receptor and disrupt receptor–ligand interactions. Alternatively, SNPs in intracellular domains of the receptor may change the kinetics of receptor–G protein interaction, alter phosphorylation of the receptor, disrupt the binding of accessory proteins necessary for scaffolding, or interefere with the internalization and desensitization of the receptor. Each of these problems would result in a diminished signal downstream of the receptor, leading to altered, and possibly inappropriate, cellular response to stimuli.
Most SNPs are synonymous or non-coding and do not change protein structure. It is believed that there is selection against non-synonymous polymorphisms in most genes because of the high likelihood of a deleterious effect on protein structure and function. Other forms of genetic variation include promoter polymorphisms and variable number of tandem repeats (VNTRs). The functional consequences of these genetic changes have been more difficult to deduce. Each may have an indirect impact on receptor expression by altering the efficiency of transcription or translation. Diminished or enhanced receptor expression will influence the signaling cascade downstream of neurotransmitter, thus altering the overall cellular response to the stimulus.
SNP frequency is known to vary by gender and by race (36, 37) . There are many reports of enriched frequencies of SNPs in African populations (36, 38) . This ethnic variability––diminished diversity in non-African populations––is consistent with a genetic bottleneck during migration out of Africa and should be carefully considered when choosing a population for association analysis. SNPs that occur in genes located on the X or Y chromosomes may produce a dose-dependent effect in males and females. The single copy of the X or Y chromosome in men may impact the frequency at which these SNPs occur among males. Additionally, the X-chromosome undergoes the phenomenon of X-chromosome inactivation (XCI)––the silencing of one copy of the X-chromosome in females. XCI does not change the genotypic frequency of a polymorphism, but it may alter the phenotypic consequence. By means similar to XCI, several autosomal genes are imprinted––an event wherein one or several genes on a chromosome are selectively inactivated in a tissue- or developmental-specific manner. Polymorphisms that occur in an imprinted gene may have no phenotypic consequence in that individual, yielding a greater degree of complexity to association studies. Lastly, some genes, including the 5-HT2A receptor gene (39) , are reported to be imprinted in a polymorphic manner, suggesting that the gene is silenced in some individuals but not in others.
SNPs within the 5-HT2A Receptor Gene
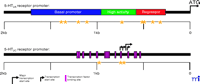
The serotonin 5-HT2A receptor gene lies on chromosome 13q14–q21 and contains two introns, with the coding region of the gene spanning 1.4 kb. At present, there are nine known polymorphisms within the promoter region of the 5-HT2A receptor (Table 1⇓ and Figure 3⇓). A functional map of the promoter region (40) makes predictions about the consequences of polymorphisms possible; however, direct functional analyses are needed to validate these predictions. The only functional study of an SNP within the promoter examined the -1438A/G polymorphism using a luciferase reporter gene assay (41) . These studies showed no difference between the A and G alleles, and no further studies have been conducted to elucidate the impact of this or any other polymorphism within the 5-HT2A regulatory region. There have, however, been several association studies performed comparing these polymorphisms, with the majority of these studies examining the occurrence of the -1438A/G polymorphism (Table 2⇓).
Frequency and Distribution of SNPs
Association Studies with SNPs of the Human 5-HT2A and 5-HT2C Receptors
Distribution of promoter SNPs and functional map of regulatory regions for the 5-HT2A/2C receptor genes. Asterisks indicate the location of SNPs within the promoter. SNPs occur within all known regulatory regions and may interfere with binding of various transcription factors.
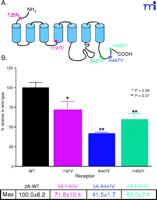
To date, there have been seven polymorphisms discovered within the coding region of the 5-HT2A receptor gene (Table 1⇑); of these, five are non-synonymous. Although discoveries of four of the non-synonymous coding SNPs were published several years ago, little has been done to investigate the impact of these amino acid substitutions on receptor function. The first report, examining platelets from individuals heterozygous for the SNP encoding H452Y, revealed alterations in platelet 5-HT2A receptor function including a blunting of the serotonin-induced Ca2+ mobilization curve (42) . Although the authors speculate that the Tyr452 receptor variant exists in a partially desensitized state, these initial results merit further investigation. For example, it is possible that the Tyr452 receptor is not partially desensitized but rather has altered kinetics of G protein coupling. Functional analyses in cells transfected with the variant receptor are needed to fully characterize the consequences of the tyrosine allele on receptor function. One other published study on the functional impact of these non-synonymous SNPs was performed in SF9 cells, which lack endogenous Gq (43) . Because a non-native system was used, the end-point was not Gq activation but G16 activation––a promiscuous G protein that has not been previously demonstrated to couple effectively to the 5-HT2A receptors; thus, the physiological relevance of these findings is unclear. Research is currently underway in our laboratory to better understand the functional consequence of these 5-HT2A receptor non-synonymous SNPs. Analyses of PLC activation reveal a diminished signal downstream of each receptor variant (Figure 4⇓). Preliminary evidence suggests that the molecular mechanism underlying this common phenotype (e.g., reduced signaling) may be distinct for each variant. Despite the scarcity of functional data, several groups have attempted to correlate specific SNPs of the 5-HT2A receptor with a disease state (Table 2⇑). The common SNP 102T/C, which occurs within the population at a frequency of about 50%, is the most widely studied variant in association studies, with nearly thirty reports scrutinizing the role of this SNP in schizophrenia. Recent meta-analysis of these data suggests that the C-allele predicts modest, but significant, increased risk for schizophrenia (44). This SNP is in complete linkage disequilibrium with the -1438 A/G SNP.
Non-synonymous 5-HT2A R SNPs. A. Distribution of SNPs in the 5-HT2AR. Asterisks indicate sites of amino acid changes. B. Maximum PI hydrolysis response of NIH3T3 cells stably expressing 5-HT2AR variants; cells were treated with varying concentrations of 5-HT for 30 minutes to generate full dose-response curves. Data represent the maximum response obtained following drug treatment (n=5).
SNPs within the 5-HT2C Receptor Gene
The 5-HT2C receptor gene contains three introns and is located at Xq24; the coding region spans approximately 1.4 kb (45) . To date, there are three known SNPs in the promoter region of this receptor, one non-synonymous SNP in the coding region, and one SNP in the 3′ untranslated region (Table 1⇑ and Figure 3⇑). There have been two reports of dinucleotide repeats (GT and CT) in the promoter region of the 5-HT2C receptor (46, 47) . Although the locations of the GT repeats appear to differ, this reflects the numbering system that was used by the two groups; the two GT repeats are in fact identical. Functional characteristics of the promoter SNPs in the 5-HT2C receptor have been examined in only two reports, both of which evaluated haplotypes using luciferase reporter gene constructs (46, 48) . Yuan’s group found that promoter activity of two of the haplotypes, including positions -1,027 and -697, had higher activity than the wild-type haplotype. Many groups have begun to examine promoter haplotypes in disease association studies as summarized in Table 2⇑. Of these, the most promising seems to be the -759C/T polymorphism. This SNP, along with haplotypes containing other nucleotide substitutions in the promoter, is associated with increased transcription levels of the 5-HT2C receptor and may confer resistance to obesity and Type II diabetes (46) . Together, these studies support the hypothesis that SNPs in the promoter region of the 5-HT2C receptor may regulate levels of the receptor protein, which could potentially change the neuronal regulation of many physiological processes including food intake. Therefore, continued identification and characterization of genetic variation in the promoter region of this receptor are important.
Only one SNP has been identified in the coding region of the 5-HT2C receptor, converting a cysteine (Cys) to a serine (Ser) at amino acid codon 23 in the first hydrophobic region of the receptor. The group that originally discovered this SNP has conducted the only functional work done on this polymorphism. Recombinant human 5-HT2C-Cys and 5-HT2C-Ser receptors were expressed in frog oocytes, and ability of 5-HT to regulate Ca2+ -activated chloride channels was evaluated (49) . Although no significant differences were found, a more detailed characterization of this variant in mammalian cell lines is needed because so many association studies have found positive correlations with the 5-HT2C-Ser allele (Table 2⇑). Given the prominence of RNA editing of the 5-HT2C receptor in human brain (18, 24) , it is also important to examine the polymorphic variants in different edited backgrounds.
Numerous human studies have examined the association of the Cys23Ser SNP in the 5-HT2C receptor with disease states and drug responses (Table 2⇑). The most frequently examined phenotype is clozapine response in schizophrenic patients; both positive and negative results have been reported, with more evidence supporting a lack of association. The Cys23Ser polymorphism is also associated with visual hallucinations and hyperphagia in Alzheimer’s patients (50) ; however, this finding has not been confirmed. Association studies of 5-HT2C receptor polymorphisms require greater care to control for epigenetic factors such as XCI.
Future Directions
Genetic association studies of SNPs in the 5-HT2A and 5-HT2C receptor genes are largely inconclusive. In addition to population stratification, there are several other possible explanations for the discrepancies. Many of the disease states that may be influenced by the serotonergic system (e.g., schizophrenia and major depressive illness) are polygenic in nature and are likely affected by environmental factors. Furthermore, as suggested by Lerer and Macciodi (51) , conflicting association studies may be indicative of a strong association with a subclass or a sub-phenotype of the illness. Thus, there is a need for a large, well-defined disease population with several sub-phenotypes that can each be specifically evaluated with respect to the SNP of interest. Such thorough investigations of the intricacies of brain diseases are more likely to uncover the role of SNPs in the 5-HT2A and 5-HT2C receptors. Similar arguments apply to human studies of potential abnormalities in RNA editing of the 5-HT2C receptor, which have so far included only small patient samples analyzed by a laborious subcloning strategy. Establishment of a high-throughput assay of the pattern of RNA editing would advance both human as well as laboratory investigations. There is also a need for a more thorough analysis into the functional consequences of the various molecular and genetic alterations in the 5-HT2A/2C receptors, so that we may better comprehend the accumulating human data. Such studies will move us closer to establishing a relationship between the biological effects of receptor alterations and the biological basis of brain disease.
- © American Society for Pharmacology and Experimental Theraputics 2003
References

Elaine Sanders-Bush, PhD , (left) is a Professor in the Departments of Pharmacology and Psychiatry, and is Director of the Neuroscience Graduate Program. She has been a member of ASPET since 1971. Lisa Hazelwood, BS , (right) and Hugh Fentress, BS , (center) are advanced graduate students in the Pharmacology graduate program at Vanderbilt, performing thesis research in the Sanders-Bush laboratory. Address all correspondence to ESB. E-mail elaine.bush{at}vanderbilt.edu; fax 615 322-4421.