Lights on for Low Oxygen: A Noninvasive Mouse Model Useful for Sensing Oxygen Deficiency
Recent advancements in clinical imaging, including magnetic resonance imagine (MRI), positron emission tomography (PET) and computed axial tomography (CAT), as well as those in molecular imaging, including bioluminescence (e.g., luciferase) and fluorescence (e.g., various fluorescent proteins) have provided the means to study complex biological processes in vivo and in real time, without compromising patients, animals, or data collection.
Optical imaging from bioluminescent or fluorescent proteins expressed in cells or tissues is the most widely used method for real time imaging. The genes encoding firefly luciferase or fluorescent proteins can either be fused with cis regulatory elements to monitor gene regulation events or within the coding sequence of a target gene to monitor protein-protein interactions. These engineered genes are then put into mice, which subsequently found a transgenic mouse line, thus allowing for organ- or organism-wide expression and study. To detect the fluorescent proteins, these reporters only need to be excited by light; however, the detection of the fluorescent proteins is restricted because of the limited penetration of the excitation light and the absorption of the emitted fluorescence. To detect the light emitted by luciferase protein, the animals are injected with luciferin, the non-light emitting substrate that after enzymatic action emits light, just prior to imaging. The advantage of luciferase is that the light emitted from deep tissues can easily be detected; however, such bioluminescence is not easily distinguished with regard to which cells or even which tissues are emitting the light. On the other hand, both bioluminescence and fluorescence in vivo can be repeatedly analyzed quantitatively in real time [see (1) for a more extensive overview].
The occurrence of hypoxic regions is a common feature of various physiological and pathophysiological conditions. Hypoxia occurs in tissue that is blocked from the blood supply resulting in a lack of nutrients, oxygen, and ischemia. In addition, actively growing tumors develop hypoxic sites because blood flow to the center of these tumors is often restricted (2, 3). The hypoxic stimulus provokes adaptive processes on multiple levels. Individual cells redistribute their metabolic enzyme composition towards anaerobic metabolism, and the amounts of glycolytic enzymes and glucose transporters in hypoxic cells and tissues are increased. Neovascularization, as indicated by the increased expression of vascular endothelial growth factor, occurs in the region close to the hypoxic tissue, whereas erythropoiesis, marked by the induction of erythropoietin (EPO) expression, is an organism-wide effect.
Many of the responses to hypoxia are facilitated by the induction of a major transcription factor: the hypoxia-inducible factor (HIF) (4, 5). HIF is a heterodimer consisting of the stable, constitutively expressed aryl hydrocarbon receptor nuclear translocator (ARNT) and an unstable, regulated α-subunit. There are three isoforms of the α-subunit. HIF-1α and HIF-2α are activators of transcription, whereas HIF-3α appears to act as a negative regulator of HIF activity (6, 7). The predominant isoform, HIF-1α, is ubiquitously expressed and its role in the physiological response to hypoxia was characterized over a decade ago (8).
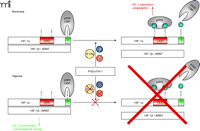
Detrimental scenarios such as hypoxia require an immediate response. Consequently, the regulation of the transcription factor involved in regulating these assaults must be finely tuned and instantaneous. Because HIF-1α is constitutively expressed, activation is quickly obtained by posttranslational modification. Under conditions that include physiological concentrations of oxygen, HIF-1α is hydroxylated at two proline residues (402 and 564) by specific prolyl-4-hydroxylases (PHD), which are members of the EGLN (egg-laying nine homologous) protein family (9, 10). Hydroxylation at these prolines allows the E3 ubiquitin ligase pVHL (von Hippel Lindau protein) to bind and to ubiquitinate HIF-1α and thereby mark it for proteasomal degradation, thus keeping the HIF-1α protein amounts low. The PHDs are the oxygen sensors for HIF-1α regulation, requiring dioxygen for the hydroxylation of HIF-1α. A decrease in oxygen levels results in a loss of PHD activity and an accumulation of non-hydroxylated, non-ubiquitinated HIF-1α. The stabilization of HIF-1α allows dimerization with ARNT, which elicits its transcriptional activity (Figure 1⇓). More details of the PHD-mediated regulation of HIF-1 have been discussed in recent reviews (11–14).
The involvement of HIF-1α in various disease states (Table 1⇓) makes it a prominent target for drug design studies (15–17). The ability to assess the effects drugs have on HIF-1α protein amounts or on HIF-1 activity, therefore, represents an interesting tool for drug design studies. Indeed, to do so in vivo would be extremely useful for evaluating the tissue and target specificity of the individual drug or event.
Safran et al. (18) have reasoned that because the HIF-1 PHDs are the key oxygen sensors and can respond immediately to a decrease in oxygen levels, that a reporter of PHD activity would be valuable for monitoring oxygen availability and would facilitate the development of pro- or anti-angiogenic compounds. To this end they engineered a mouse model that expresses the oxygen-dependent degradation (ODD) domain of HIF-1α fused to the firefly luciferase gene. Fusing the ODD domain to luciferase renders the luciferase protein sensitive to oxygen-dependent hydroxylation and subsequent ubiquitin-dependent degradation under normal physiological circumstances. To ensure the widespread expression of the fusion protein, the ODD-Luc cDNA was integrated into the ROSA26 locus, which is expressed in most tissues.
In cell culture, the ODD-Luc fusion protein is responsive to hypoxia and hypoxia mimetics. In vivo, in normal oxygen concentrations, the ROSA26/ODD-Luc mice emitted significantly less light then mice that express wild-type luciferase under the control of the same ROSA26 promoter, indicating that the fusion protein was sensitive to ODD-mediated degradation. The light emitted from the ROSA26/ODD-Luc mice was detected in the region of the kidneys, which is consistent with their finding that mammals breathing room air possess hypoxic zones in their kidneys (18). When the ROSA26/ODD-Luc mice were placed in low oxygen, the emission of light increased 5- to 10-fold relative to mice breathing room air. Previously, detection of PHD activity or stabilization of HIF-1 under similar conditions had not been possible.
Preclinical studies indicate that increasing HIF-1 activity might be beneficial to prevent tissue hypoxia that arises from myocardial infarctions or stroke (19). The ROSA26/ODD-Luc mice are ideally suited for identifying and characterizing small molecule inhibitors of PHD that would increase HIF-1 activity. Indeed, Safran et al. showed that treatment of animals with FG-4383, an inhibitor of the PHDs, resulted in a rapid and dose-dependent increase in bioluminescence from these mice (18). FG-4383-induced bioluminescence was greatest in the regions of the kidney. In agreement with the luciferase results, treatment of ROSA26/ODD-Luc mice with FG-4383 increased HIF-1-mediated expression of EPO mRNA in the kidneys and circulating concentrations of EPO protein in the blood.
Safran et al. concluded that, unlike traditional invasive methods of harvesting tissue or serum, the ROSA26/ODD-Luc mice can provide a rapid non-invasive measurement of possible agonists of HIF-1 activity (18). Furthermore, given that multiple time and dose determinants can be measured with the same animal, the ROSA26/ODD-Luc mice are useful for generating spatial and temporal information. The ROSA26/ODD-Luc should also be useful for monitoring hypoxic regions of solid tumors of cancer-prone mouse strains. In this case the bioluminescence would be used to report increases in HIF-1α which are associated with increased angiogenesis and a poor prognosis.
The ROSA26/ODD-Luc mice are a valuable addition to the rapidly growing list of genetically engineered mice designed to report biological functions. The ability of these mice to rapidly report, in a non-invasive manner, the effects of potential drugs capable of regulating these functions, opens up a new and exciting future for identifying and characterizing these compounds in vivo.
(Patho-) Physiological Situations Associated with Increased HIF-1 Activity
Oxygen concentration-dependent regulation of HIF-1α. Under normoxic conditions (and in the presence of 2-OG and Fe2+)HIF-1α is hydroxylated at two proline residues by specific prolyl-4-hydroxylases (PHDs) which allows the E3 ubiquitin ligase pVHL to bind to HIF-1α and mark it for proteasomal degradation. In addition, HIF-1α is regulated by the aspargine hydroxylase factor inhibiting HIF-1 (FIH-1) in the C-terminal transactivation domain (C-TAD), which inhibits p300/CBP binding to HIF-1α (20). If oxygen concentrations decrease, the PHDs and FIH-1 are inactivated because of the limited oxygen supply. This inactivation allows HIF-1α to accumulate, form the active transcription factor HIF-1 with the α-subunit, recruit transcriptional cofactors, and initiate the transcription of responsive genes. HIF-1, hypoxia-inducible factor; pVHL, von Hippel Lindau protein; ARNT, aryl hydrocarbon receptor nuclear translocator; ODD, oxygen-dependent degradation domain; 2-OG, 2-oxoglutarate.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
- © American Society for Pharmacology and Experimental Theraputics 2006
References

Matthew R. Young, PhD, is a Staff Scientist in the Laboratory of Cancer Prevention at the National Cancer Institute, Frederick, Maryland. He received his PhD in Biochemistry from the University of Maryland School of Medicine. After a postdoctoral fellowship at ABL-Basic Research Program at the NCI-Frederick, he joined Nancy Colburn’s lab. In 2000 he was promoted to Staff Scientist in the LCP where he is working on developing mouse models engineered to image the early events of carcinogenesis. E-mail: Youngm{at}ncifcrf.gov; fax 301 846-6907.

Tobias Schmid, PhD, is a postdoctoral fellow in the Laboratory of Cancer Prevention (LCP) at the National Cancer Institute, Frederick, Maryland. He received his PhD from the University of Kaiserslautern, Germany, in 2004, where he was working on hypoxia-induced signaling mechanisms. In 2005 he joined, the LCP where he is working on the transformation repressor Pdcd4. E-mail: Tschmid{at}ncifcrf.gov; fax 301 846-6907.