Event
September 11-13, 2006 Robinson College, Cambridge, United Kingdom
eGenomics 2006
eGenomics III: Cataloguing our complete genome collection
George Garrity discusses NamesforLife and PhenBank at Cambridge. He will also chair Monday’s second session: “Databases and Metadata capture and Exchange efforts”.
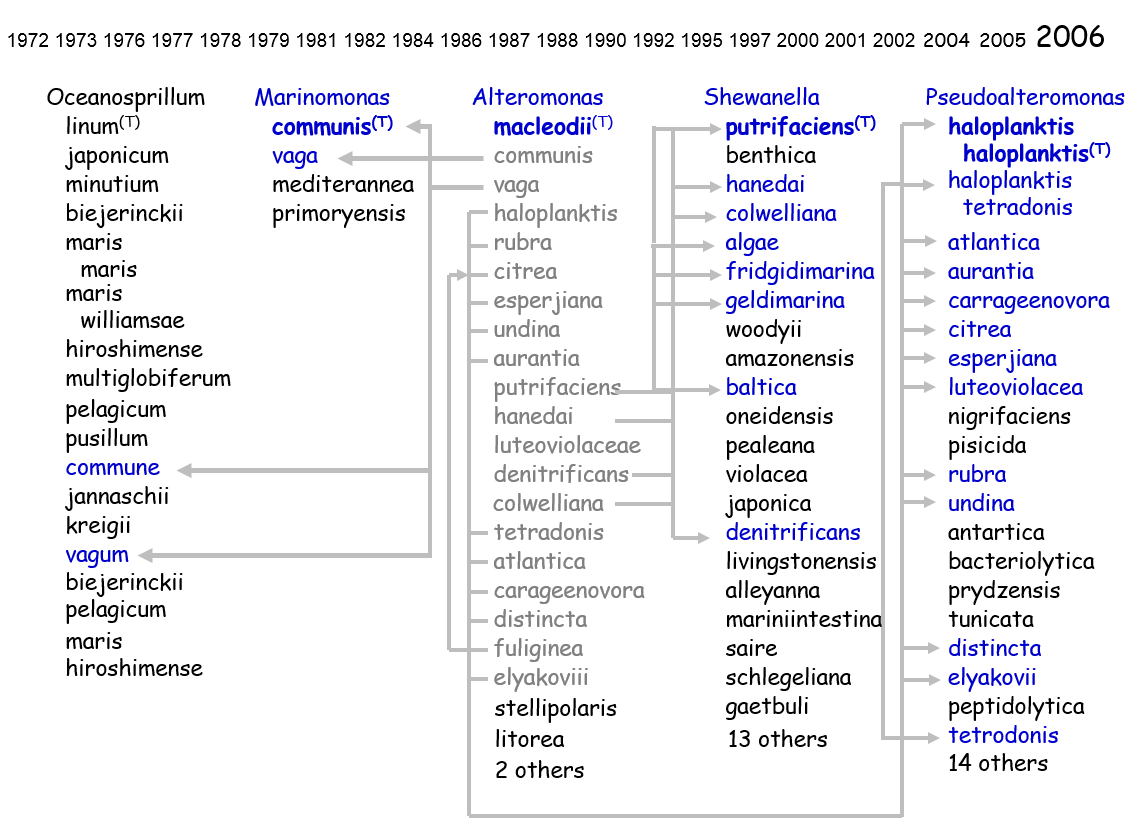
Names, taxon concepts and exemplars are independent. Names are fixed in time and are bibliographic events, tied to a particular published description. The taxon concept, however, drifts once it comes into usage, as non-type exemplars are added to the global sample set. There is also a critical need to always tie the data (phenotype and genotype) to the correct source strain.
When one looks at the environmental data, it becomes difficult to accurately interpret results across studies, especially when one is dealing with survey data comprised of a single measurement (e.g., a 16S rRNA sequence). One of the reasons is that investigators use their own identifier to label the data (and strains). More importantly, many of these labels are not unique.
We are in the process of updating our prototype to identify all of the high quality 16S rRNA sequences that have come from type strains held in different Biological Resource Collections (BRCs).
We have been using heatmaps of evolutionary distance matrices to visualize sequence similarity and to uncover annotation errors in the 16S rRNA sequence data set for about five years. Last year, we published the SOSCC algorithm which can undertake this process in an automated manner.
What is particularly useful is that the method allows us to examine 1,000–10,000 sequences simultaneously, thereby revealing the otherwise hidden structure associated with more distant taxonomic relationships.
Download Presentation (2.8MB PowerPoint)
[permalink] Posted September 7, 2006.